













| General Information: |
|
| Name(s) found: |
XRCC1-PA /
FBpp0070730
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 614 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
single strand break repair
[IEA]
base-excision repair, AP site formation [IMP] nucleotide-excision repair [ISS][NAS] response to DNA damage stimulus [IMP] |
| Molecular Function: |
damaged DNA binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPIALFKSVR EVSSEDAVHV AANLLKENAG KKWRTKAPGE KSAYVVLEFE EPQQITGIDI 60
61 GNEHAAFIEV LVSRTGCQAD DFRELLLSSS FMTPIESKNS SNPNRVRCFS STSLAESALP 120
121 EKWKLLQIVC TQPFNRHVPY GLSFVKVHVV AAAAPKVKSL VPQGVMQFGT FKLREESPDS 180
181 ETDNQVNRFH SWKKQTQHKS PAISATSTAA AIRDAGSTAL RRLSAGKVSS LAASPKTSTS 240
241 PSPVLNALRS VSPTPLDAKP LDRNRASLLF GDDDEDVKDA EVNAKKQRLS KHLEADKDRR 300
301 RLEQEKERES RKKKSSRHSL DKSISKEDKP PAKEHNSSSR EKSTAKEEKE QLRKEERSHS 360
361 KEEKSRRKEK SKHDHRSSDT SNSKESSRPT AQKRHSSPTA VAVTPAKKQK IVDTAIQYRP 420
421 LNQLLKGVVL VISGIQNPDR ADLRSKAVAL GAKYKADWGS GCTHLICAFK NTPKYNQVKG 480
481 KGKIVTRSWI EKCYELKKYL PWRRYALDTN DIGQPESDEE LCDEATRPRR DSSKDDVAML 540
541 EDSKLTALAD QNDVEMNPDA LSGIDTEDEL QRVAEENKTK KAKLRTTVKT KSQDIYEVST 600
601 DEEDYLELKK KQID |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
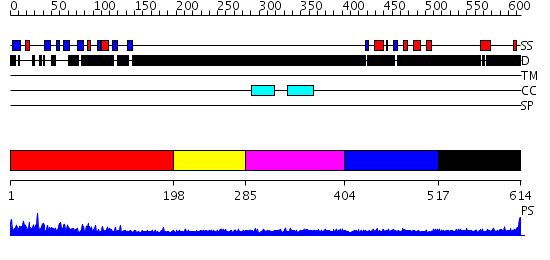
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 42.69897 | N-terminal domain of xrcc1 | |
| 2 | View Details | [198..284] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [285..403] | 1.14 | No description for 2j63A was found. | |
| 4 | View Details | [404..516] | 9.522879 | No description for 2d8mA was found. | |
| 5 | View Details | [517..614] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)