













| General Information: |
|
| Name(s) found: |
norpA-PE /
FBpp0290690
[FlyBase]
norpA-PA / FBpp0070619 [FlyBase] norpA-PB / FBpp0070618 [FlyBase] |
| Description(s) found:
Found 41 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1095 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA]
inaD signaling complex [IPI][TAS] rhabdomere [IDA] |
| Biological Process: |
diacylglycerol biosynthetic process
[TAS]
phospholipid biosynthetic process [TAS] phosphatidylinositol metabolic process [IMP] light-induced release of internally sequestered calcium ion [TAS] adult locomotory behavior [IMP] rhodopsin mediated phototransduction [IMP] phospholipid metabolic process [TAS] mucosal immune response [IMP] phototransduction [IMP][NAS][TAS] response to abiotic stimulus [IMP] intracellular signaling pathway [IEA] entrainment of circadian clock [IMP] negative regulation of compound eye retinal cell programmed cell death [IMP] elevation of cytosolic calcium ion concentration involved in G-protein signaling coupled to IP3 second messenger [TAS] sensory perception of smell [IMP] phosphoinositide metabolic process [IMP] calcium-mediated signaling [TAS] deactivation of rhodopsin mediated signaling [IDA] |
| Molecular Function: |
phosphoinositide phospholipase C activity
[IEA][NAS][TAS]
phospholipase C activity [TAS] calcium ion binding [IEA] GTPase activator activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKKYEFDWI IPVPPELTTG CVFDRWFENE KETKENDFER DALFKVDEYG FFLYWKSEGR 60
61 DGDVIELCQV SDIRAGGTPK DPKILDKVTK KNGTNIPELD KRSLTICSNT DYINITYHHV 120
121 ICPDAATAKS WQKNLRLITH NNRATNVCPR VNLMKHWMRL SYCVEKSGKI PVKTLAKTFA 180
181 SGKTEKLVYT CIKDAGLPDD KNATMTKEQF TFDKFYALYH KVCPRNDIEE LFTSITKGKQ 240
241 DFISLEQFIQ FMNDKQRDPR MNEILYPLYE EKRCTEIIND YELDEEKKKN VQMSLDGFKR 300
301 YLMSDENAPV FLDRLDFYME MDQPLAHYYI NSSHNTYLSG RQIGGKSSVE MYRQTLLAGC 360
361 RCVELDCWNG KGEDEEPIVT HGHAYCTEIL FKDCIQAIAD CAFVSSEYPV ILSFENHCNR 420
421 AQQYKLAKYC DDFFGDLLLK EPLPDHPLDP GLPLPPPCKL KRKILIKNKR MKPEVEKVEL 480
481 ELWLKGELKT DDDPEEDASA GKPPEAAAAP APAPEAAAAA EGAAEGGGGA EAEAAAANYS 540
541 GSTTNVHPWL SSMVNYAQPI KFQGFDKAIE KNIAHNMSSF AESAGMNYLK QSSIDFVNYN 600
601 KRQMSRIYPK GTRADSSNYM PQVFWNAGCQ MVSLNFQSSD LPMQLNQGKF EYNGGCGYLL 660
661 KPDFMRRADK DFDPFADAPV DGVIAAQCSV KVIAGQFLSD KKVGTYVEVD MFGLPSDTVK 720
721 KEFRTRLVAN NGLNPVYNED PFVFRKVVLP DLAVLRFGVY EESGKILGQR ILPLDGLQAG 780
781 YRHVSLRTEA NFPMSLPMLF VNIELKIYVP DGFEDFMAML SDPRGFAGAA KQQNEQMKAL 840
841 GIEEQSGGAA RDAGKAKEEE KKEPPLVFEP VTLESLRQEK GFQKVGKKQI KELDTLRKKH 900
901 AKERTSVQKT QNAAIDKLIK GKSKDDIRND ANIKNSINDQ TKQWTDMIAR HRKEEWDMLR 960
961 QHVQDSQDAM KALMLTVQAA QIKQLEDRHA RDIKDLNAKQ AKMSADTAKE VQNDKTLKTK1020
1021 NEKDRRLREK RQNNVKRFME EKKQIGVKQG RAMEKLKLAH SKQIEEFSTD VQKLMDMYKI1080
1081 EEEAYKTQGK TEFYA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
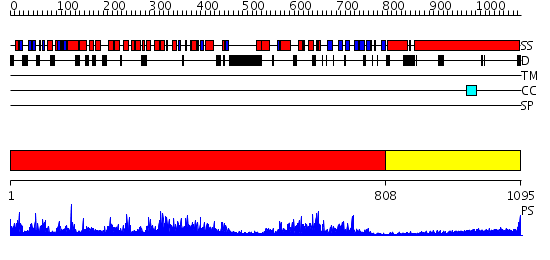
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..807] | 1000.0 | No description for 2fjuB was found. | |
| 2 | View Details | [808..1095] | 23.522879 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)