













| General Information: |
|
| Name(s) found: |
CG10260-PB /
FBpp0070427
[FlyBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2178 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
phosphoinositide-mediated signaling
[IEA]
phosphoinositide phosphorylation [IEA] phosphorylation [NAS] |
| Molecular Function: |
1-phosphatidylinositol 4-kinase activity
[ISS][NAS]
binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTMTASDKYT YQRTVLCLAR VLAGIQPTPW DKVHVHLATH STSHSASPPG NRLFALTLSG 60
61 GHKTDMRGRS GGGGKAVQTL FRYCPQENAA GVFCLDTRAQ DAVIALGIYF LEGGCQHEGQ 120
121 IVPYLLRLAK CLPKAVWIDD ARSNKVERVR IPSAEKFSFC LNTLLSDIAA KCPDSREEII 180
181 LNQVETLSAL ANIVKSSRDS SSAPPPIILC KATVPLLFGL ARSMGRYASN DPPLLCRIFP 240
241 PELLPIQKGG GRDGTGSSSS ASGTCGGSFS SSERLAATHH FRPIIPRSMS GSLAQAQNAS 300
301 YDDGRQRCAG GKPSKPSLHS YFSVPYDPRT HFFTRYGSSF NQFPNMRVCE SPTKGGPRPL 360
361 YRVPPFPIQH LQTIFAVSKK LLTKDTLEHL DEQASDIFSL HQIKGYCYKS FSETLNLVLV 420
421 TLLRELLQHQ VDLPTPFTKD VQEFVKRLFL NGQTELQNKQ QDQERERREE NGIAVVNKYK 480
481 VNVMANAACV DLLVWAIRDE TEADKLCGRL SQKLNLELSH KIVMDHMPLL MVCLEGLGKL 540
541 AHKFPNIAGT SISYLRDFLV APSPILGKLH DHAMQSLAQQ KKEKELTPFK IAVQHSDSRT 600
601 AVVIYGDNQK PPGSGTGRSG HAAFESLRDA AIENLSIALR AAHTLDQFCV PALVANVSNR 660
661 LFTAEKSGSE SQGECHKSNL VSLNIIVMLG HVAVALKDTS KTTQNILQFF IQRFCKVPSE 720
721 QNALIVDQLG CMIISQCETH VFDEIMKMFS RVTVQSASLA YTSDPEHRKQ FHHVSDAVVN 780
781 ALGNIAANIQ GDAEMLELLG KLLELFVQIG LDGERSYDNT PGAQKASSRA GNLGMLIPVI 840
841 AVLVRRLPPI KNPRQRLHKL FKDFWAYCVV MGFTNARLWP ADWYQGVQQI AAKSPLLISQ 900
901 TAHKSDMREL NYTLAIKSDS VNELRSQILV LLEHSSDNVA TAINKLSFAQ CTYLLSVYWL 960
961 EMLRVENADE PSLEPIMSYL CDTALQRDKT GIWQCVKCVA DQVFEKFRNV LYAHDEIREK1020
1021 VLESQATLLL VYFNHIHKPI QMVADQYLSF LVDRFPHLLW NRRVLWCMLD ILQLLAYSLS1080
1081 LDPNEETPTL RVVSTPYTLQ LMDSLPAREL RLKDFADRCQ GIVNEAMKWA PRSTRSHLQE1140
1141 YPNQIPTPVL AHHSGLALAF DSVVSSSAQH TGTMSKRPSC VNSDTPRFVS VLCLRSKYAG1200
1201 EISGLLSVLS EKDKAGLADR LVSDVWEACA EKSDARHRGA LWRATAYLII CSEISRKLLH1260
1261 AVASSQLELF TESAMETAVE CWQWVLTARQ DLELCFIQEM VSAWQTTFEK RMGLFAWETE1320
1321 VTHPLAAYEG CKLVSKPILI APHLIWLQLL SEMVDTAKYC NRDKVEMFCL LLHRCLPVLK1380
1381 SSKQNRQVST VGCRFKLLQC GLSLLQGNTI PKSLSRNILR ERIYSNALDY FCGPPTCPNQ1440
1441 SREQLLEDIM ILLKFWQTMR SEKKHLVTSE VGDYDLTNAS VSSTQMLAVR NNPETASLIS1500
1501 GGGLVNDYTR SMSASGNAVG MGMGVAGGGS SSGWYNTIPH STSTLSKRSN RSKRLQYQKD1560
1561 SYDKDYMKKR NLILELLAVE LEFLITWYNP NSLPDLIVPG EEQITEWRNR PYKSTVWRDY1620
1621 ARLAWCYNPA LAVFLPQRIK NAEIIDEEVS RLVCSDPIAV CHIPEALKYL CTTKNLLQES1680
1681 PDLVYILSWS PVTPIQALAY FSRQYPSHPL TAQYAVKTLS SYPAESVLPY IPQLVQALRH1740
1741 DTMGYVVEFI KNISRRSQIV AHQLIWNMQT NMYMDEDQQH KDPNLYEALD QLSQSIIASF1800
1801 SGAAKRFYER EFDFFGKITA VSGEIRSFAK GIERKNACLA ALSRIKVQGG CYLPSNPEAM1860
1861 VLDIDYSSGT PMQSAAKAPY LARFRVYRCG ITELETRAME VSNNPNSQED AKMTLGVESW1920
1921 QAAIFKVGDD VRQDMLALQV ITIFKNIFQQ VGLDLFLFPY RVVATAPGCG VIECVPNAKS1980
1981 RDQLGRQTDS GLSEYFQHQY GDESSKEFQA ARANFVKSMA AYSLIGYLLQ IKDRHNGNIM2040
2041 IDKDGHIIHI DFGFMFESSP GGNIGFEPDM KLTDEMVMIM GGKMDSPAFK WFCELCVQAF2100
2101 LAVRPYQDAI VSLVSLMLDT GLPCFRGQTI NLLKQRFVAT KNNKEAAAHM LAVIRNSYQN2160
2161 FRTRTYDMIQ YYQNQIPY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
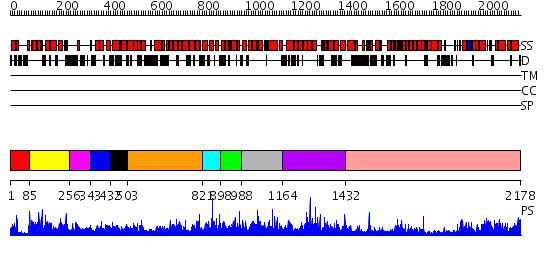
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [85..255] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [256..342] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [343..431] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [432..502] | 1.029986 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [503..820] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [821..897] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [898..987] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [988..1163] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1164..1431] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1432..2178] | 125.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)