













| General Information: |
|
| Name(s) found: |
FBpp0305122
[FlyBase]
FBpp0301695 [FlyBase] FBpp0301694 [FlyBase] FBpp0301693 [FlyBase] CG42666-PD / FBpp0291795 [FlyBase] |
| Description(s) found:
Found 42 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 742 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
exonuclease activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSCNKNSSS RITTTNNNVT TTSNISGNSS NKRCPQKYTG NTLAVNSNML AYQQQATAGG 60
61 KNSTRKSRHN NNKQNHQQQQ QQQQQQFDHI HKRNGTAGGT GSECSPAAPT SNWLQRGYGR 120
121 QFVHSSCIED KLLGHHNPGH RNSTSSDATS EDIDSSSELP NKSKGNSCAG SIHASGGSKS 180
181 RSKRHQKQSG QQTRNPDHNG QRTGAATPCT GQKAKQGNAK WRQQQPNPSS NNQQQQMQQQ 240
241 MQQQKRGRNK KPGKWSNNII DNPNSNHGGS SQQKTCDSSS SSALSSPTSS ATTSATSSPT 300
301 NSPTSYPRAN GNRKRIEAAL EWRREQELGR QNANFLVPPL RQYEQLEFDD KTVVQQLRRH 360
361 IIDNHLLRVY GFPVDSVVHE GAIEIFKCMP HMSFAAALAE TAQAVTVINY HDGLTNRAEE 420
421 EVANSTDSGN SSPQSLDSES GGEWSGSECE SSNSSSSSAG EAALGLELKY CHFVPVERSL 480
481 SKPCARCKRH FLVDELTGQY LTHEECQYHS GKYQRYYDGV SHGRWTCCNE GEESSPGCCL 540
541 GDRHVWSGSV VGVNGPYYDF VRTEHRGSGE DEPAVYALDC EMSYTGRGLD VTKVSLVALN 600
601 GQLVYEHFVR PVCDIIDYNT QYSGITETDL CSGAKSLAEV QRDLLQLITA DTILIGHGLE 660
661 NDLRALRLVH NTLIDTSISF PHCNGFPYRR ALRHLTKVHL KRDIQAGDGT TGHSSFEDSR 720
721 ACMELMLWRV NRELDPAWSW DD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
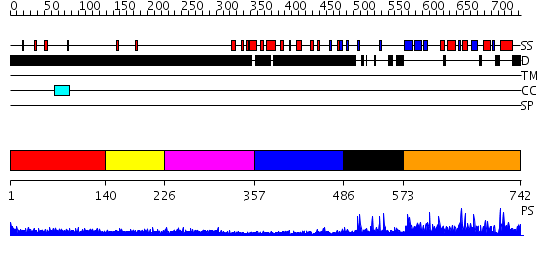
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | 4.201996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [140..225] | 2.226991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [226..356] | 2.230996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [357..485] | 2.202995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [486..572] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [573..742] | 46.0 | human ISG20 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)