













| General Information: |
|
| Name(s) found: |
CG11403-PA /
FBpp0070235
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 861 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nucleotide-excision repair
[ISS]
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IEA] |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] helicase activity [ISS] ATP-dependent DNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQYTPNQRL DTPGAQEFGF PYSPYEIQEQ LMQELFQVLE RGQVGIFESP TGTGKSLTLT 60
61 CGALTWLARH EELVRTEMLA RIRGVEQELA KLKEESEQSS NWLESQGKSR AQRAELLRLQ 120
121 HLQALLDKQE QQLDQIRKGA KKHKRQGRVH PGKLEELEKD LDPESDSDSE HETADGPQEA 180
181 AEDRYRPVQI FFCSRTHSQL AQIVAELRKT PHGQSVRSIS LGSRQQLCGN PAVRKLKHVG 240
241 LMNERCLDMA TKKARPNPSK KSRLTAEANS RCPFKAASLV ESLRDLALTE PLDIEELANE 300
301 GTACGGCSYY ASRSAVEHAQ LILLPYQLLL QKSARNQLGI SLKGSIVIVD EAHNLLDSVA 360
361 QLHGSEISRQ QLERAKVQIS GYKDHFQKRF TTKNLLKINQ IIFIIRRLLK ILDQRKELQS 420
421 NGCSMMRTYE LTAEGDFFNI DLCELLDFCA RTRFARKVQG RADRMEREPR PSENQAPVST 480
481 ARSLILQRLA SEQKLKEKTK SVKRRVEDIN KEDKAEELQE QQKPTKKPVE EVAPSPIRPL 540
541 LAFLETLTSN AEDGRILVDP VGGTLKYILL DPAEQFADIV AEARAIVIAG GTMQPTKELK 600
601 EQLFTGCHDR LVERFYNHVV ANDAILPFVI SNGPSGAPLS FKFAHRGSAE MLRELSMILR 660
661 NLCQVVPGGV VCFLPSYEYL DKVYKYLEQS GTLETISGRK SVFREVSGSA EQLLDNYALA 720
721 IKRPASYGAL LLSVVGGKLS EGLNFADDLG RAVLVVGLPY SNSLSPELRQ RMQHLDEKLG 780
781 PGAGNEYYEN LCVKAVNQCI GRAVRHIKDY ACVYLLDKRF ADPKIRGKLP KWISRHIVEA 840
841 NAANGGFGAV QGRTARFFKG R |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
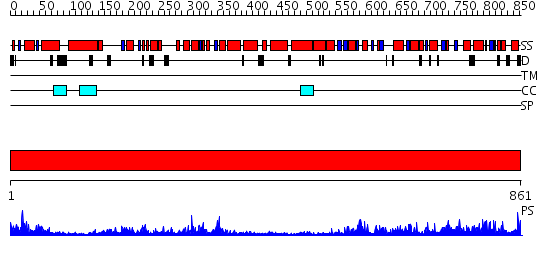
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..861] | 1.43 | No description for 2p6rA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)