













| General Information: |
|
| Name(s) found: |
FBpp0311396
[FlyBase]
FBpp0305856 [FlyBase] Thd1-PA / FBpp0088175 [FlyBase] |
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1738 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nucleotide-excision repair
[NAS]
mismatch repair [IDA] response to DNA damage stimulus [IMP] |
| Molecular Function: |
pyrimidine-specific mismatch base pair DNA N-glycosylase activity
[IDA][NAS]
double-stranded DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQEESGSTPL LSSSFSYTPT VAPLVLKIAL IQAARSFAAS KYTIKVLLSL NLALSEQAIV 60
61 QKAVKPICTV MASEVDASSG PEDGTVPTLM SLTPYITNLE HGNNESGKYV SGLPNNRKRQ 120
121 KLSILEHNTI QNNDVDNTEV EPNMDPKSKM SDTKSNKYSE MEKHLNDNSK IVIEGTISIG 180
181 NSKRKSPEKL VPYDNDCYMP PEQALVTSVE LVVPAPQTHS SNTPGRQSEE PNLSTLGESS 240
241 TTPSSTIDNK LQYSTAGLYN STSSTSILAN DKIVGCANDS SNLNLRIPTK LVVTTASGDI 300
301 LIDDRRASLW TPHHDESGQR QQRTGTASSD TKQEPMNVSS ELSYHHQNRH SELLLQIEKE 360
361 SSGSFLQASP IPLQDNHNNA ASGQFGQTEE TSNIDSQSHN NFYAQMMQPQ HLLHNQHQQS 420
421 MHEHSPRHQQ PASYSGYITH YQNPPMFGAH QSEHHQRLNQ QQQPLQHLLD CHGHLEQSTP 480
481 ISQQNQHHLS QQIHQHQHQQ THQRLPLREN YHDIIMDDFH EEPSHAFKLT LSPSNTKPEN 540
541 QDDGYETSAG DVLTPNSHSS STHSITPQHQ MQHSNIVLMT QNQKKSDDLQ LTKVTLSGEA 600
601 HTDPNACSSN SSQGQVLASQ SHLELSEGTR CSSHASVVDP YSFMGEELHM HSPSHRHLDA 660
661 VTTGPGRYGI LVSNDTPECL SREMYRHSQQ STTVLEQTDS SSCGINFKPM PKKRGRKKKL 720
721 VAVNADTSQM TTPVDQQKVS AGRADCEDGG GDQAAKPKER KKHDRFNGMS EEEVIKRTIP 780
781 DHLCDNLDIV IVGINPGLFA AYKGHHYAGP GNHFWKCLYL AGLTQEQMSA DEDHKLIKQG 840
841 IGFTNMVARA TKGSADLTRK EIKEGSRILL EKLQRFRPKV AVFNGKLIFE VFSGKKEFHF 900
901 GRQPDRVDGT DTFIWVMPSS SARCAQLPRA ADKVPFYAAL KKFRDFLNGQ IPHIDESECV 960
961 FTDQRIRLCS AQQQVDIVGK INKTHQPPLG DHPSSLTVVS NCSGPIAGDA ECGIVAEESD1020
1021 QVQSEKMIPQ MDPTVPSSSN ATDGKSFSYT AENTPLLPVS NHNPSINENN YLSVMGSQQP1080
1081 LSQQPLEKKK RGRPKKIKGQ DIIDHSVGGK ASIAGQHIPS HDFNNILNLS VMSGGGTIET1140
1141 PKKKRGRPKK LKPAIDNIMT VKQLQHGNNN LNTTAGLSAS SMHPISMEHI AASPQSSHQM1200
1201 PPSLYNTPPP SHLLYTASAS PMASPALNCN YTQVHGHGTP PVGQVASVAQ GSSPVIDTQN1260
1261 DHLAQQKQSH HGNLGAGLDM RDHPHLGETP PPSSPNMCST VDFDPPDEHS GSQVGSRVQN1320
1321 KAVELDHQHP QIMEKVQYDS PVPNTEANPA HPHENYQQWL SPHPHQSNQP AQKLTHRQQH1380
1381 PPMHHFHQEQ TENWQRYEEQ NSNPYMVISA HHQHLSPRLG NQTHQNSSPS GHISSDVAHK1440
1441 SLCGLESLVD QIPAIREQDC SNIPLATVAA AAAAVESRIL SLQHQHQHPL QPHQQNQQNQ1500
1501 QQQLKQCKQE NSAHRESCRP TSENSNVSNS NFSVSSLAAS ASSARTDNAI YGNGETKGNN1560
1561 ESSHHNSCDT NIDYPIHNQS AYHHTPHLIG SALGTNVNNS EPNLHTISHP HPPHPHPHSM1620
1621 YVDQAHHMAH IPSVNVNSMY GPAYGSHPQH TTGEYPGTHG HYSLGGSVQT AVPTSSATLH1680
1681 VPSPNYPFGH HPYGHTPPQA NYPSYTHPHT HHHHSHPSHH LTVFDHLKPS DISGYGGF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
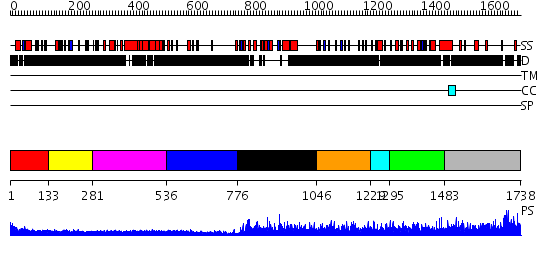
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [133..280] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [281..535] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [536..775] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [776..1045] | 93.69897 | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | |
| 6 | View Details | [1046..1228] | 1.060984 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1229..1294] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1295..1482] | 2.070996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1483..1738] | 0.95 | No description for 2jrnA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)