













| General Information: |
|
| Name(s) found: |
FBpp0311451
[FlyBase]
Hcf-PA / FBpp0088195 [FlyBase] Hcf-PB / FBpp0088194 [FlyBase] |
| Description(s) found:
Found 59 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1500 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
H4/H2A histone acetyltransferase complex [IDA] |
| Biological Process: |
regulation of histone acetylation
[IDA]
|
| Molecular Function: |
transcription factor activity
[ISS]
transcription coactivator activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGSDFVDPA FSSGERISAS DLNSEHIIQA ENHSFANRIS MDMDVPDGHQ LDSNLTGFRW 60
61 KRVLNPTGPQ PRPRHGHRAI NIKELMVVFG GGNEGIVDEL HVYNTVTNQW YVPVLKGDVP 120
121 NGCAAYGFVV EGTRMFVFGG MIEYGKYSNE LYELQATKWE WRKMYPESPD SGLSPCPRLG 180
181 HSFTMVGEKI FLFGGLANES DDPKNNIPKY LNDLYILDTR GVHSHNGKWI VPKTYGDSPP 240
241 PRESHTGISF ATKSNGNLNL LIYGGMSGCR LGDLWLLETD SMTWSKPKTS GEAPLPRSLH 300
301 SSTMIGNKMY VFGGWVPLVI NDSKSTTERE WKCTNTLAVL DLETMTWENV TLDTVEENVP 360
361 RARAGHCAVG IQSRLYVWSG RDGYRKAWNN QVCCKDLWYL EVSKPLYAVK VALVRASTHA 420
421 LELSWTATTF AAAYVLQIQK IEQPLNTSSK LLSNNIVQQG TPTSAETSGI NISANRSGSA 480
481 LGLGVEATST VLKLEKESLQ LSGCQPETNV QPSVNDLLQS MSQPSSPASR ADKDPLSSGG 540
541 GTTFNLSTSV ASVHPQISVI SSTAAVTGND TASPSGAINS ILQKFRPVVT AVRTSTTTAV 600
601 SIATSTSDPL SVRVPSTMSA NVVLSSSSST LRIVPSVTAS HSLRIASSQA SGNNCRSSSA 660
661 INILKTALPN VAVQSQPTSS TTTSIGGKQY FIQKPLTLAP NVQLQFVKTS GGMTVQTLPK 720
721 VNFTASKGTP PHGISIANPH LASGITQIQG STVPGSQIQK PIVSGNVLKL VSPHTMAGGK 780
781 LIMKNSNILQ MGKVTPNVMG GKPAFVITNK QGTPLGNQQI IIVTTGGNVR SVPTSTVMTS 840
841 AGGSASGTNI VSIVNSTSTT PSPLQALSGQ KTLISNQSGV KMLRNISSVQ ASSSMAFGQK 900
901 QSGTPIHQKT ALYIGGKAVT VMSTNTSMAA SGNKVMVLPG TSSNNSPATT TALSARKSFV 960
961 FNAGGSPRTV TLATKSINAK SIPQSQPVTE TNNHSVATIK DTDPMDDIIE QLDGAGDLLK1020
1021 LSESEGQHGS EENENNGENA TSSSASALFT GGDTAGPSRA QNPIVMEHPV DIIEDVSGVS1080
1081 STTDVNETAI VSGDTIESLK MSEKENDDVK SMGEKSILSD DCHQPTTSET EAATILTTIK1140
1141 SAEALVLETA EIRKDHTGCT IGSLKENQDE NKKFKQRQES SPSQNIHQFQ NVDGSQLEAL1200
1201 ASAALLQAAT SDTTALALKE LIERPESETN TRSSNIAEIQ QNNVQSTLAV VVPNTSQNEN1260
1261 QKWHTVGVFK DLSHTVTSYI DSNCISDSFF DGIDVDNLPD FSKFPRTNLE PGTAYRFRLS1320
1321 AINSCGRGEW GEISSFKTCL PGFPGAPSAI KISKDVKEGA HLTWEPPPAQ KTKEIIEYSV1380
1381 YLAVKPTAKD KALSTPQLAF VRVYVGAANQ CTVPNASLSN AHVDCSNKPA IIFRIAARNQ1440
1441 KGYGPATQVR WLQDPAAAKQ HTPTVTPNLK RGPEKSTIGS SNIANTFCSP HKRGRNGLHD1500
1501 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
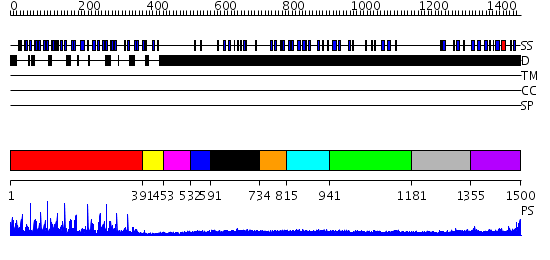
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..390] | 48.0 | Crystal structure of the Kelch domain of human Keap1 | |
| 2 | View Details | [391..452] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [453..531] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [532..590] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [591..733] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [734..814] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [815..940] | 1.189986 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [941..1180] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1181..1354] | 1.03 | No description for 1zlgA was found. | |
| 10 | View Details | [1355..1500] | 6.045757 | Solution structure of C-terminal fibronectin type III domain of mouse 1700129L13Rik protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)