













| General Information: |
|
| Name(s) found: |
Vha100-1-PG /
FBpp0084746
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 850 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar proton-transporting V-type ATPase, V0 domain
[ISS]
synapse [IDA] |
| Biological Process: |
ATP synthesis coupled proton transport
[IEA]
|
| Molecular Function: |
calmodulin binding
[IDA]
hydrogen-exporting ATPase activity, phosphorylative mechanism [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSLFRSEEM ALCQLFLQSE AAYACVSELG ELGLVQFRDL NPDVNAFQRK FVNEVRRCDE 60
61 MERKLRYLEK EIKKDGIPML DTGESPEAPQ PREMIDLEAT FEKLENELRE VNQNAEALKR 120
121 NFLELTELKH ILRKTQVFFD EQEGGVNQTT ESMTRALITD EARTAGASMG PVQLGYMEKS 180
181 NEREDYLPCF VAGVILRERL PAFERMLWRA CRGNVFLRQA MIETPLEDPT NGDQVHKSVF 240
241 IIFFQGDQLK TRVKKICEGF RATLYPCPEA PADRREMAMG VMTRIEDLNT VLGQTQDHRH 300
301 RVLVAAAKNL KNWFVKVRKI KAIYHTLNLF NLDVTQKCLI AECWVPLLDI ETIQLALRRG 360
361 TERSGSSVPP ILNRMQTFEN PPTYNRTNKF TKAFQALIDA YGVASYREMN PAPYTIITFP 420
421 FLFAVMFGDL GHGAIMALFG LWMIRKEKGL AAQKTDNEIW NIFFGGRYII FLMGVFSMYT 480
481 GLIYNDIFSK SLNIFGSHWH LSYNKSTVME NKFLQLSPKG DYEGAPYPFG MDPIWQVAGA 540
541 NKIIFHNAYK MKISIIFGVI HMIFGVVMSW HNHTYFRNRI SLLYEFIPQL VFLLLLFFYM 600
601 VLLMFIKWIK FAATNDKPYS EACAPSILIT FIDMVLFNTP KPPPENCETY MFMGQHFIQV 660
661 LFVLVAVGCI PVMLLAKPLL IMQARKQANV QPIAGATSDA EAGGVSNSGS HGGGGGHEEE 720
721 EELSEIFIHQ SIHTIEYVLG SVSHTASYLR LWALSLAHAQ LAEVLWTMVL SIGLKQEGPV 780
781 GGIVLTCVFA FWAILTVGIL VLMEGLSAFL HTLRLHWVEF QSKFYKGQGY AFQPFSFDAI 840
841 IENGAAAAEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
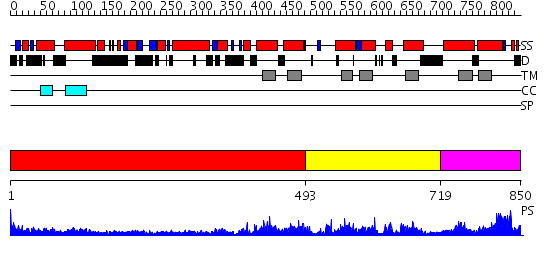
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..492] | 5.69897 | COLICIN IA | |
| 2 | View Details | [493..718] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [719..850] | 3.112969 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|