













| General Information: |
|
| Name(s) found: |
beta4GalT7-PA /
FBpp0084130
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 322 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[ISS]
Golgi apparatus [IDA] |
| Biological Process: |
chondroitin sulfate biosynthetic process
[NAS]
carbohydrate metabolic process [IEA] heparan sulfate proteoglycan biosynthetic process [NAS] glycosaminoglycan biosynthetic process [NAS] proteoglycan biosynthetic process [ISS] |
| Molecular Function: |
metal ion binding
[ISS]
galactosyltransferase activity [IDA][NAS] xylosylprotein 4-beta-galactosyltransferase activity [IDA][ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVNISTINWV FVCGLSFCLG GIAVLSLMPL GSDCVCPLSN PLAKLAGGGE GVSVIKKEPA 60
61 DEKQPQPHDH GASVHKMALL VPFRDRFEEL LQFVPHMTAF LKRQGVAHHI FVLNQVDRFR 120
121 FNRASLINVG FQFASDVYDY IAMHDVDLLP LNDNLLYEYP SSLGPLHIAG PKLHPKYHYD 180
181 NFVGGILLVR REHFKQMNGM SNQYWGWGLE DDEFFVRIRD AGLQVTRPQN IKTGTNDTFS 240
241 HIHNRYHRKR DTQKCFNQKE MTRKRDHKTG LDNVKYKILK VHEMLIDQVP VTILNILLDC 300
301 DVNKTPWCDC SGTAAAASAV QT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
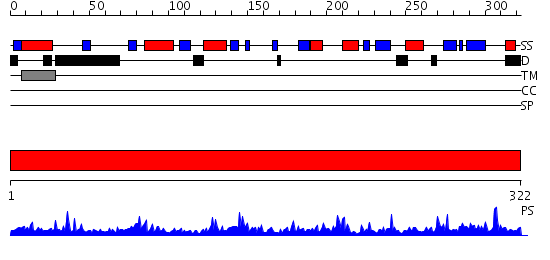
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..322] | 90.69897 | beta 1,4 galactosyltransferase (b4GalT1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|