













| General Information: |
|
| Name(s) found: |
Dys-PB /
FBpp0083180
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1669 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoskeleton
[ISS]
dystrophin-associated glycoprotein complex [IPI][ISS][NAS] |
| Biological Process: |
muscle organ development
[ISS]
muscle cell homeostasis [IMP] neuromuscular synaptic transmission [IDA] establishment of cell polarity [IMP] imaginal disc-derived wing vein morphogenesis [IMP] imaginal disc-derived wing vein specification [IMP] regulation of short-term neuronal synaptic plasticity [IDA] |
| Molecular Function: |
zinc ion binding
[IEA]
structural constituent of muscle [ISS] actin binding [ISS] calcium ion binding [IEA] structural molecule activity [ISS] structural constituent of cytoskeleton [ISS] cytoskeletal protein binding [ISS] WW domain binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAKPPPPIP PTLGGDDSGT HGPKLAPEIP RINSELAAQA ALRGQQLLRK QGSQEQHATN 60
61 TLPAHKTGAP PPLPPTQSRP VPAIPARGAS DRAAPPEIPP KRHSLKSMPD GQPMMVPGLP 120
121 PTMRQPPPLP RKPASTQSSA QNSAQSSPLA GMKFKDKPPP PPEKHSTLSA EGMAARRCSN 180
181 PFEMPPPPPP LVLQSAVAAL SEQSSKNGLN PVPSPAPTRA SEAKKINQRS IASPTEEFGS 240
241 EDALRGIESG LRNMERAMQE QMNLRNMEAA VQNNFDLSFK ASLAAGARHL GGGSVNLRTA 300
301 NYERNLSLDE GRAGDSRQSL EELKQLRAQA QQQSLLNNSS SSSSNSQVEQ SMRSTIEHHM 360
361 RSLDRNLPLE LQYSRHRFQN NLNAVAAAGG GGGNSSTGNA VANSGTSGSQ QPPMPLSSEF 420
421 REQIRQQLLG NIPPQVVHNM GQSPGSAAAA ALLQHQAQSR AAAAAAAAAA AAAAAQVAAG 480
481 SGALSREELR MRRRSSHDET QLTQNSSGGI QVTRLREHWD ETSQCVLQRA AQLKNMLSDS 540
541 QRFEAKRLEL EKWLARMEQR AERMGTIATT ADILEAQQKE QKSFHAELHQ NKQHFDIFNE 600
601 LTQKLIAVYP NDDTTRIKKM TEVINQRYAN LNSGVINRGK QLHAAVHSLQ SFDRAMDQFL 660
661 AFLSETETLC ENAESDIERN PLMFKDLQSE IETHRVVYDR LDGTGRKLLG SLTSQEDAVM 720
721 LQRRLDEMNQ RWNNLKSKSI AIRNRLESNS EHWNALLLSL RELTEWVIRK DTELSTLGLG 780
781 PVRGDAVSLQ KQLDDHKAFR RQLEDKRPIV ESNLTSGRQY IANEAAVSDT SDTEANHDSD 840
841 SRYMSAEEQS RELTRSIRRE VGKLSEQWNN LIDRSDNWKH RLDEYMTKMR QFQKILEDLS 900
901 SRVALAEQTK TSWLPPSSVG EANEQMQQLQ RLRDKMTTAS ALLDDCNEQQ SFFTANQVLV 960
961 PTPCLSKLED LNTRMKLLQI AMDERQKVLC QAGAQQTHEN GDDGRTTSNS GTIGPLPNLG1020
1021 QSVKPPWERA TTAANVPYYI DHERETTHWD HPEMIELMKG LADLNEIRFS AYRTAMKLRS1080
1081 VQKRLALDRI SMSTACESFD RHGLRAQNDK LIDIPDMTTV LHSLYVTIDK IDLTLMLDLA1140
1141 INWILNVYDS QRTGQIRVLS FKVGLVLLCK GHLEEKYRYL FRLVADTDRR ADQRRLGLLL1200
1201 HDCIQVPRQL GEVAAFGGSN IEPSVRSCLE QAGISQEAID GNQDISIELQ HFLGWLQHEP1260
1261 QSLVWLPVLH RLAAAEAAKH QAKCNICKEY PIVGFRYRCL KCFNFDMCQK CFFFGRNAKN1320
1321 HKLTHPMHEY CTTTTSTEDV RDFTRALKNK FKSRKYFKKH PRVGYLPVQS VLEGDALESP1380
1381 APSPQHTTHQ LQNDMHSRLE MYASRLAQVE YGGTGSNSTP DSDDEHQLIA QYCQALPGTS1440
1441 NGSAPKSPVQ VMAAMDAEQR EELEAIIRDL EEENANLQAE YQQLCSKEQS GMPEDSNGMQ1500
1501 HSSSSMTGLS GQGEQGQDMM AEAKLLRQHK GRLEARMQIL EDHNRQLEAQ LQRLRQLLDE1560
1561 PNGGGSSATS SGLPSAPGSA LNSKPNTLQT RSVTASQLNT DSPAKMNQQN GHYEHNSKNS1620
1621 SGLVTVITEQ ELESINDDLE DSSSSNTTNT TTTTTTTATT EKTCVELQK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
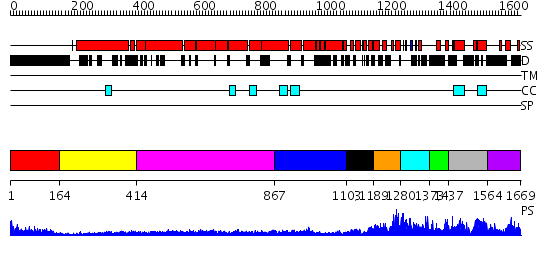
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..163] | 6.09691 | No description for 1y0fA was found. | |
| 2 | View Details | [164..413] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [414..866] | 17.39794 | Heavy meromyosin subfragment | |
| 4 | View Details | [867..1102] | 6.39794 | Crystal Structure of Repeats 15, 16 and 17 of Chicken Brain Alpha Spectrin | |
| 5 | View Details | [1103..1188] | 5.30103 | No description for 2dfsA was found. | |
| 6 | View Details | [1189..1279] | 64.69897 | Dystrophin | |
| 7 | View Details | [1280..1372] | 26.045757 | No description for 2e5rA was found. | |
| 8 | View Details | [1373..1436] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1437..1563] | 2.02 | No description for 2fxmA was found. | |
| 10 | View Details | [1564..1669] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)