













| General Information: |
|
| Name(s) found: |
CG5316-PB /
FBpp0083174
[FlyBase]
CG5316-PC / FBpp0083172 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 662 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
precatalytic spliceosome [IDA] |
| Biological Process: |
single strand break repair
[ISS]
nuclear mRNA splicing, via spliceosome [IC] |
| Molecular Function: |
zinc ion binding
[IEA]
catalytic activity [IEA] damaged DNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSWSSALIKD ISKPENLIIS SEIAVVIADK FPKAQHHYLV LPLADIPSIF HLNRSHLSLL 60
61 EELHLLARNV VEVKGVRWQD FNVGFHAEPS MQRLHLHVIS KDFVSTSLKT KKHWNSFNTE 120
121 LFVPYTKLYA QLEKENSISR LPKSLKDELL AKPLICNQCE FVARNLPSLK GHLVGHLQDP 180
181 KSVCQRVRLG NQFFPTAGYR TSELAYCFDF VDFYEYKKQM EVDKLAYIRD ELQRKLNDKR 240
241 NFLIESDRAV VMKADYPKSQ YHFRVVAKEE FRDITQLTEA QLPLLDHMMD LANQIIEKQK 300
301 HLESRNFLIG FKVNTFWNRL NLHVISNDFY SMAMKRISHW NSFNTELFMP FQIAYMMLSV 360
361 QGSIESISEE TYNNLQEKTP LRCNQCEFVT NMLLDLKAHL YQHWQRKEDE RDQKKKVDKI 420
421 IQMISETKLD EAEAKPKLLN EEEPIQAQPV AAIAQYPNEH LGKPLTPQQQ PGKQQAQNVY 480
481 DKNINGPSVN MMNQNNPNNP FRNTPHLNRQ SQKPPHPRSG PRGPMAPWTG PRFPCHQQQN 540
541 RFRPPGFNAC RQPYPPYHSG HQQFPNASSV GGGQTGLPGQ GQGPRPKWNS NKIFNQQNRQ 600
601 NTVQAQPQAQ NQQTNQQQIQ NSNKNQTPKK KPWKNRLQPV GKVQNQGGAN RDPAPPSNSK 660
661 PS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
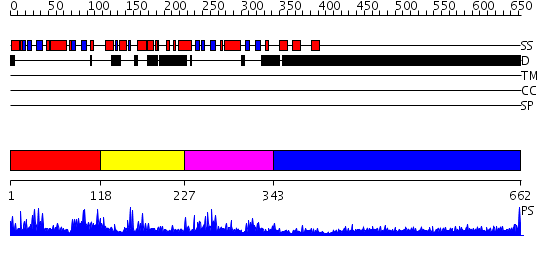
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 29.221849 | Protein kinase C inhibitor-1, PKCI-1 | |
| 2 | View Details | [118..226] | 3.56 | No description for 2yt9A was found. | |
| 3 | View Details | [227..342] | 24.522879 | HIT family hydrolase from Clostridium thermocellum Cth-393 | |
| 4 | View Details | [343..662] | 7.522879 | No description for 1y0fA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)