













| General Information: |
|
| Name(s) found: |
gi|7300435
[NCBI NR]
gi|24648048 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1612 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
MAPKKK cascade
[IDA]
response to oxidative stress [IMP] protein amino acid phosphorylation [IDA][NAS][TAS] mucosal immune response [IMP] response to stress [IDA][IMP] |
| Molecular Function: |
ATP binding
[IEA]
receptor signaling protein serine/threonine kinase activity [NAS] MAP kinase kinase kinase activity [IDA][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNRRRVRTI DYLALQQSLR LQKTPAATTN AEEQVAREEE NGNGHHSTVT AETPPTPPIP 60
61 PIPPIRLRRE QSVEEDVASK DSIDPRRESA VAYRGSVIQP PGQSSHLIAQ EEVCKMRRKK 120
121 VEYRVKQTPS RPVQMTRNRI GALEEDMPPE DELAAHYEAF GTTPPRTRLK IKNRDWERKQ 180
181 KVVNVTASAD LPASVGGTPK KTRTARSRVL RRNTMDCALL NEMFVNDESK RTDKRLQSLL 240
241 RDSEREMKNS LATTAVAAPR GNSFHETAHP LESLDQIMPL NSEAMAPIPL RIASKVVESC 300
301 NRYRCVSSRP IGYRSSAPPL AFAAQLPAGM LRSDGRATPG LGKRKDFHET FANLIKLGSV 360
361 DRQDAKLSQE EHTWQTELKD LIWLELQAWQ ADRTVEQQDK YLFEARQGVS DLLTHIINYK 420
421 FQPRYRREPS LISLDSGTHS DSNSNASSPL PSKMCQGCMS LYCKDCMDHQ ELALREVEGM 480
481 LTRLEAAEAL YPSSQAMGAL HPIYKSQSFV GRIKSMCLWY NITKQNKLKL SILGKILARL 540
541 QDEKFSWPVC TSSYIATDSG SSSASGVEND DSAVNSMDSS KPPSMAGSAS RKGVTPCHKV 600
601 QFMLNDATHV PGETSSSNES TSTEVSQWSS ECSHSHMRKG SMHDINIFSV EPLGTCSSNG 660
661 TGEQSTGLYR KFIENVLKSR GLAKSLAFLH KLHNVALYKA HIALEKPGAE DFDYESDAES 720
721 IEEDVPRLDP EISREQVVEL RTYGYWSEEA QSINLPSYIP TFVFLSGIPL QFMHEFLRMR 780
781 LETRPVRPNP LSLEQLMKEL REGLTLALTH RERYQRHITT ALVENEAELG SYISILNHYD 840
841 ATVRKTFELY LEYIDQLVLV AVPEGNQKSV LEKEWMFTKL ISPMIKGMHT LASQKFCGII 900
901 SKLLRSISER LVKRTVELDQ QIDGTADTDD NEEVKWQLLT ICRETQSLLT VERERSIKVL 960
961 FFAKTFCRDV ETTDFHREHY EHDVANQQHD FICSDVKAAF KLLQQDVLQV RNKLTAIIEG1020
1021 VQKRCCLSNM RDLDEQDKQA VLSRTREILH QGYKFGFEYH KDVIRLFEQK IMDQKDSGAH1080
1081 TVDLALGIIA YAKMWMHFVM ERCERGRGMR PRWASQGLEF LILACDPQIT QHLDDDEFEA1140
1141 LKQQMDRCIS HVIGITSEPE KVAKKKASPR TRKTSSPATS RSRTPTRTPM SAGMVLNPNT1200
1201 PPLQSPPYNK LLHPQFSLKE DVSGTSYSPV DSSDYVDTPC QRSANGELRL LVPQTPPTPA1260
1261 SPGKSSLEST PLALRQERVR DAVNRLDMDL EDGLRERRLI GQVKSLNSSD KVHIRARSVH1320
1321 FRWHRGIKIG QGRFGKVYTA VNNNTGELMA MKEIAIQPGE TRALKNVAEE LKILEGIKHK1380
1381 NLVRYYGIEV HREELLIFME LCSEGTLESL VELTGNLPEA LTRRFTAQLL SGVSELHKHG1440
1441 IVHRDIKTAN IFLVDGSNSL KLGDFGSAVK IQAHTTVPGE LQGYVGTQAY MAPEVFTKTN1500
1501 SDGHGRAADI WSVGCVVVEM ASGKRPWAQF DSNFQIMFKV GMGEKPQAPE SLSQEGHDFI1560
1561 DHCLQHDPKR RLTAVELLEH NFCKYGRDEC SSEQLQMQVR GSFRRNVATS SS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
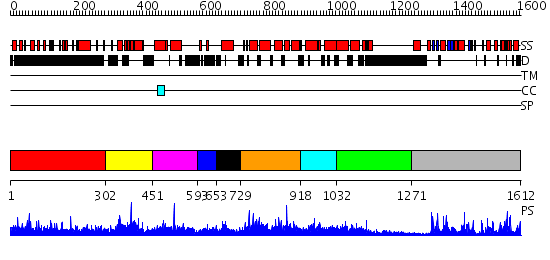
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..301] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [302..450] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [451..592] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [593..652] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [653..728] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [729..917] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [918..1031] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1032..1270] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1271..1612] | 78.0 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)