













| General Information: |
|
| Name(s) found: |
ps-PO /
FBpp0291833
[FlyBase]
ps-PN / FBpp0291832 [FlyBase] ps-PH / FBpp0110328 [FlyBase] ps-PG / FBpp0110327 [FlyBase] ps-PF / FBpp0110326 [FlyBase] |
| Description(s) found:
Found 62 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 572 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IEP]
|
| Molecular Function: |
mRNA binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEDATMETC PSPETGDSRK RPLDSDPENE QTKRSHFSSG ESVCSGIEVE IENNNNNHIH 60
61 HGETTYHMKI LVPAVASGAI IGKGGETIAS LQKDTGARVK MSKSHDFYPG TTERVCLITG 120
121 STEAIMVVME FIMDKIREKP DLTNKIVDTD SKQTQERDKQ VKILVPNSTA GMIIGKGGAF 180
181 IKQIKEESGS YVQISQKPTD VSLQERCITI IGDKENNKNA CKMILSKIVE DPQSGTCLNV 240
241 SYADVSGPVA NFNPTGSPYA TNQNAINSST ASLNSTLGTT IGGANSAASL LVNGTGINLS 300
301 INLGSPNPAP NLAVATQLLE HIKVAMRGSG YSETVTNEVV AALSVLAKYG VLGMGVGVSH 360
361 TNGAHSTLGN FLGVTTLDQQ TAAAASAATA SNVFGAVGQV NLEQYAAAVA SAAAASRPTQ 420
421 SQLDAAAVQF DPFRHLGSAT APAATPVSLN NNSFGLTATT GTATTAQLGG LSKSPTPGDL 480
481 SSKDSKNVEV PEVIIGAILG PSGRSLVEIQ HVSGANVQIS KKGIFAPGTR NRIVTITGQP 540
541 SAIAKAQYLI EQKINEEETK RARQIPLTTV VN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
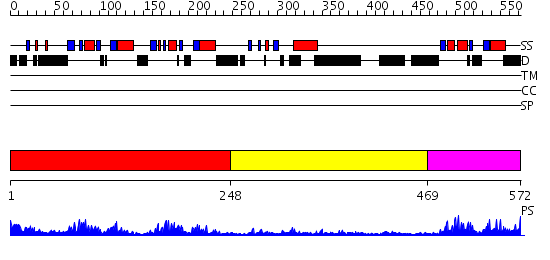
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..247] | 33.522879 | No description for 2annA was found. | |
| 2 | View Details | [248..468] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [469..572] | 17.69897 | Neuro-oncological ventral antigen 2, nova-2, KH3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)