













| General Information: |
|
| Name(s) found: |
ird1-PA /
FBpp0081466
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1342 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular response to amino acid starvation
[IMP]
positive regulation of antibacterial peptide biosynthetic process [IMP] protein amino acid phosphorylation [IEA] macroautophagy [IMP] regulation of antimicrobial peptide production [IMP] |
| Molecular Function: |
ATP binding
[IEA]
protein serine/threonine kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNQLVGIAP SQIYAVEHYF SGQFGSEIAF SSNMGSTRFF KVAKAKTDEG QIVVKVFVKH 60
61 DPTLPLEDHK ERLEGIKKTL SLANAVNCLP FQRVELIDKA AYIMREYVKH SLYDRVSTRP 120
121 FLTVLEKKWI TFQILCALNQ CHKQKICHGD IKLENILITS WNWILLSDFA SFKPTYLPED 180
181 NPADYTYFFD TSRRRTCYIA PERFVKTLAS DDDGGNGNMS VIHTDSIIRL APSYAGNTLL 240
241 PAMDIFSAGC ALLELWTEGT APFELSQLLA YRRGERDLVE KHLAGIENER LRNLLASMID 300
301 IHSMNRKSAE DYLDQERGQL FPEYFYSFLQ SYLQMFSSTP IMSPDDKIQC LHKDIGHCIK 360
361 VLTQAQDLTP EEEEKEEKKD PTLCARLPPE HDGLILIITV VTSCIRGLKQ SNTKIAALEL 420
421 LQKLSKYTTS ETILDRILPY ILHLAQKSPA KVQVQAIETL TACLGMVKDI PRSDANVFPE 480
481 YILPSITDLA SETSAVIVRV AFARNIAALA KSAVYFLEET QRNAPNDMPT PRYEAELNAL 540
541 HDIVRQAVLS LLTDSQPVVK QTLMESGICD LCAFFGKEKA NDVILSHIMT FLNDEDKNLR 600
601 GAFYDNIAGV AGYVGWQASD ILVPLLQQGF TDREEFVIAK AIRAVTILIE LGHIKKPAVT 660
661 EIVQETACYL CHPNLWVRHE ICGMIATTAR NLSAIDVQCK IMPAIGAFLK APLIQVEKPH 720
721 ILLDCVHPPV PRQIFDSVLR FQDIHHFIRA LEARSRVRSQ TRQGALPQYE EMGQTLRNLF 780
781 RRLSSEGLTD LIEMQLLAMN PFLISMKHKA LQDLDDTASG NGRIVVSRKQ VACHEYPLAD 840
841 KASKMPLDNR SSEGPTLASP ASDAAITPLG AGGLAGSPAS GAVVLGVPTA TVSAATSLGE 900
901 YTMPERNCWL ERSSECRKDL ESLTAKIKSR YNVLAISQRC SYDKLVTPYP PGWRMTGTLV 960
961 AHLHEHSESV IKLASLRPHG SLFASGSIDG TVRLWDCSKL NGNQGVNKSR QVYSANTPIY1020
1021 ALAACDSGQS LAVGGKDGSM LMMRIDRNSA KMTLQQALNQ NDHKDGPVVD MHAYDQATQS1080
1081 VIVYATLYGG IVAWDTRMQH SAWRLQNELR HGVITTICAD PTGSWLATGT SGGKHICWDL1140
1141 RFRLPIAEIK HPADSWIRKV ACHPTEPSYL ISASQSNNEV SVWNIETGQR QTVLWASPVA1200
1201 ALSSASPSDP ATTCGILSGV VDGSPFILTG SSDQRIRYWD ITNPKNSSLK VPAANDNLAD1260
1261 VAFNYGARLI DGSQVIEEQI ISMASTGDSQ QLRSSTEENP RSGPDMPTAS HHDAITDLLM1320
1321 CKDKGQIYIA SASRNGVIKL WK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
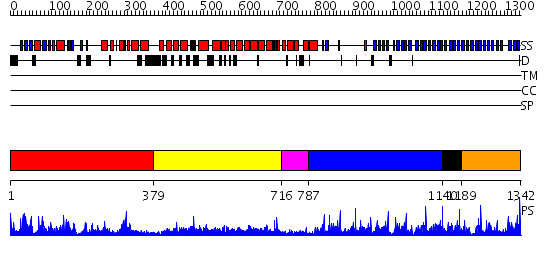
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..378] | 88.30103 | MAP kinase p38 | |
| 2 | View Details | [379..715] | 4.0 | No description for 2h4mA was found. | |
| 3 | View Details | [716..786] | 81.69897 | No description for 2ovpB was found. | |
| 4 | View Details | [787..1139] | 24.154902 | Arp2/3 complex 41 kDa subunit ARPC1 | |
| 5 | View Details | [1140..1188] | 2.3 | Actin interacting protein 1 | |
| 6 | View Details | [1189..1342] | 80.0 | No description for 2gnqA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)