













| General Information: |
|
| Name(s) found: |
p-PA /
FBpp0081373
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 826 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: |
ommochrome biosynthetic process
[IMP]
compound eye pigmentation [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADAYCLTNF IDFSLSLSLP LKHHNRIKYT CFDISDSYII FGASSGSLYL FNRNGKFLLL 60
61 IPNKHGAITS LSISANSKYV AFATQRSLIC VYAVNLSAQA TPQVIFTHLD QSVQVTCIHW 120
121 TQDEKQFYYG DSRGQVSLVL LSSFIGHSLL FNMTVHPLLY LDSPIVQIDD FEYLLLVSNC 180
181 TKCILCNTEY EDYKQIGNRP RDGAFGACFF VSPQESLQPS RIYCARPGSR VWEVDFEGEV 240
241 IQTHQFKTAL ATAPARIQRP GSGTDELDAN AELLDYQPQN LQFAKVQRLN DDFLLAFTEL 300
301 GLYIFDIRRS AVVLWSNQFE RIADCRSSGS EIFVFTQSGA LYSVQLQTLQ SHAVSLIQQS 360
361 KLLPCANLLR QHVRYFADKA REDYELKQLN PLKQLLIERQ EYELLNDISV IFDAITQCTG 420
421 SALDTHSSGG SSATTERSLS GGSSSRAPPK GVYVLENAFC DNLKQPLKTG HFKDALLTVT 480
481 GKFGKNIIKY KFNIFAEEQQ QLVRELIPAS ERSLPFKDIK ARYESGSEDQ EEEIVRRCKK 540
541 PAPQVPHISP EEKTLYNLYL IAKSAKFSRT QCVDRYRAVF DEYAAGELVN LLEKLAQVMV 600
601 EHGDTPDQAQ RNCYEMYFDY LDPEMIWEVD DATRDHIAAG FVLLNTSQNA EIVKCEHCSF 660
661 PLRFDTSCQY HELGAVLLRY FWSRGEQLKC FDVVQSVPAL LDVLAKFYLA EQNLTKVVAI 720
721 VLNYGLPELL ADVGKQLSVS AWGRCFEQFV ELQRGGRLVC ANCECISGVE QEQLGRHFFY 780
781 NWNCFLNIAL DHMSAGDTLA LIFKWSSYIP NDAIDREFYS RCLLKG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
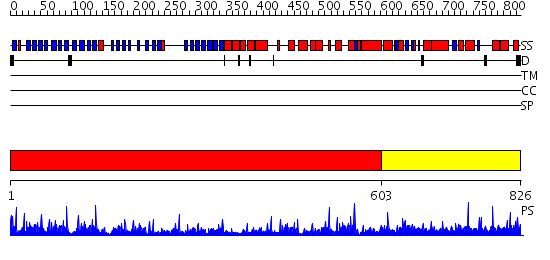
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..602] | 52.0 | Actin interacting protein 1 | |
| 2 | View Details | [603..826] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)