













| General Information: |
|
| Name(s) found: |
Hcs-PA /
FBpp0078332
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1041 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polytene chromosome
[IDA]
|
| Biological Process: |
response to heat
[IMP]
protein modification process [IEA] |
| Molecular Function: |
biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity
[ISS]
biotin-[acetyl-CoA-carboxylase] ligase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTLYYVSAT VLQSWRIQKA CSKIAEHLAQ PSSIAFYTLQ SGSDDGFDPA LASSELCCNR 60
61 NAAKVTDILW LHANQRGCCL RPLQTLHITP WISFPPAPSL LPFSYAADTL TPASTPTEAD 120
121 VPRQQRVSLS AQGEERMQLL LEADIEPLQR PSSEDTSAVR LEDYGKLIAW KIDSHLAVLI 180
181 ETDVEHFTKL LITTFLRNNL CINDQLPLLR IESVQREGDP QPFELLAKHL KRQSRISVGL 240
241 DKDGWKKHME DLRAVGVLAH QATEFEYQRN RSEGRTKSDP TTHDQRPTSE LLAKAVAVVE 300
301 PTNKAYLSPA RTTEASLKAE AKGTSTKPFP TKSDAKPATL AKSEGTPATS KEDSKLTPTK 360
361 GALKTSELEK LAAVQAAQQQ KKEAVQVSPV KAAFLAKPPM LSKHDEPDKA SDQPTKPRKS 420
421 FEAVKPLNSP PSSRAAPSRP VLQRNKDSLQ DAKPLNVLVY SDSASAREST LATLQQLLER 480
481 NVYTIYPLMP QQAAQKYWTE QTALLVVCGS VAHGIGQILV DYFLQGGKVL SLCSDILHFV 540
541 LPNYRTAEVR EHELVQFSYD KWQRVKMMHH IFCYQPSPVK KHFSTDSEES TKSHSRKPSM 600
601 ELKDLAGHSH NLDVHVLGTE ETWNTPSLML AKSLQSGGKA VFSQVHLEMN PSEFESDETK 660
661 YSILKQNERT RLEIFADLLG KYLDVQVRGG DGVDQPQPGV VYKHAYFLGR HEAKFELLEK 720
721 LRLRCSGSDN VIATPNLTMK FCGKDDKPPV ANNNVLPILI HSCPDDFSTV DYFDNLKTEH 780
781 IGRLVIYAPV VSSSMHLINN LELIHGLAVL PVQQTSGVGR RNNQWLSPPG CAMFSLQLHL 840
841 TMDSALSSRL PLLQHLVGTA IVNSLRSHEE YGVLDISIKW PNDIYANGNQ KIGGLVINTT 900
901 LQGSQAIVNI GSGINLNNSR PTVCINDLIR EYNTRVPNNK LPILKYELLI AMIFNEIERL 960
961 LGEVQNGDFD SFYALYYSLW LHSGQSVKIC LQKDQEKEAE IVGIDDFGFL EVKLPTGTIE1020
1021 IVQPDGNSFD MLKGLIIPKY Q |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
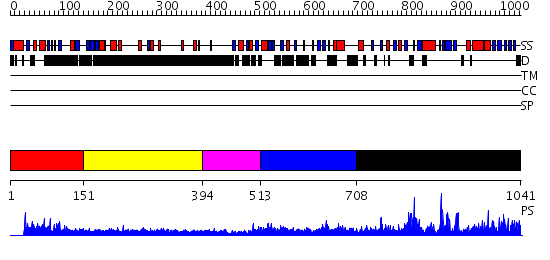
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..150] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [151..393] | 4.284997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [394..512] | 1.266994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [513..707] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [708..1041] | 85.09691 | THE E. COLI BIOTIN HOLOENZYME SYNTHETASE(SLASH)BIO REPRESSOR CRYSTAL STRUCTURE DELINEATES THE BIOTIN AND DNA-BINDING DOMAINS |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)