













| General Information: |
|
| Name(s) found: |
gek-PA /
FBpp0072194
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1637 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
intracellular signaling pathway
[IEA]
actin polymerization or depolymerization [IMP] regulation of actin polymerization or depolymerization [NAS] protein amino acid phosphorylation [IEA][ISS][NAS] |
| Molecular Function: |
ATP binding
[IEA]
small GTPase regulator activity [IEA] diacylglycerol binding [ISS] protein kinase activity [IDA] protein serine/threonine kinase activity [IEA][ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEYESSEISD ITTGSCKKRL TFLKCILSDT TSDQKWAAEF GEDTEGHQFS LDYLLDTFIV 60
61 LYDECSNSSL RREKGVSDFL KLSKPFVHIV RKLRLSRDDF DILKIIGRGA FGEVCVVQMI 120
121 STEKVYAMKI LNKWEMLKRA ETACFREERD VLVFGDRQWI TNLHYAFQDN INLYLVMDYY 180
181 CGGDLLTLLS KFEDKLPEDM AKFYITEMIL AINSIHQIRY VHRDIKPDNV LLDKRGHVRL 240
241 ADFGSCLRLD KDGTVQSNVA VGTPDYISPE ILRAMEDGKG RYGTECDWWS LGVCMYEMLY 300
301 GETPFYAESL VETYGKIMNH QNCFNLPSQE TLNYKVSETS QDLLCKLICI PENRLGQNGI 360
361 QDFMDHPWFV GIDWKNIRQG PAPYVPEVSS PTDTSNFDVD DNDVRLTDSI PPSANPAFSG 420
421 FHLPFIGFTF SLTSSSTLDS KKNQSSGFGD DTLDTISSPQ LAILPSNNSE TPVDSVQLKA 480
481 LNDQLAALKQ EKAELSKQHN EVFERLKTQD SELQDAISQR NIAMMEYSEV TEKLSELRNQ 540
541 KQKLSRQVRD KEEELDGAMQ KNDSLRNELR KSDKTRRELE LHIEDAVIEA AKEKKLREHA 600
601 EDCCRQLQME LRKGSSSVET TMPLSISSEM SSYEIERLEL QFSEKLSHQQ TRHNMELEAL 660
661 REQFSELENA NLALTKELQQ TQERLKYTQM ESITDSAETL LELKKQHDLE KSSWFEEKQR 720
721 LSSEVNLKSK SLKELQAEDD EIFKELRMKR EAITLWERQM AEIIQWVSDE KDARGYLQAL 780
781 ATKMTEELEY LKHVGTFNNN GVDNKNWRNR RSQKLDKMEL LNLQSALQRE IQAKNMISDE 840
841 LSQTRSDLIS TQKEVRDYKK RYDSILHDFQ KKETELRDLQ KGGLEYSESF LNKSTHHGLS 900
901 SAFFRDMSKN SEIIDSAESF GNESGDNFTP NFFQSGNSGM LFNYEPKYAG KNNKDHSSMK 960
961 EASVSDLSRE ESDQLVKESQ KKVPGNTAIH QFLVRTFSSP TKCNHCTSLM VGLTRQGVVC1020
1021 EICGFACHTI CCQKVPTTCP VPMDQTKRPL GIDPTRGIGT AYEGYVKVPK SGVIKRGWIR1080
1081 QFVVVCDFKL FLYDISPDRC ALPSVSVSQV LDMRDPEFSV GSVRESDVIH AAKKDVPCIF1140
1141 KIKTALIDGG LSLNTLMLAD NESEKSKWVI ALGELHRILK RNSLPNTAIF KVNEILDNTL1200
1201 SLIRNALCSV IIYPNQILLG TEDGLFYINL DQYEIARIGE SKKILQLWYI EEEQILVILC1260
1261 GKQRNLRLLP IRALEASDVE WIKVVESKNC ISACTGIIRR FPNIVYSFII ALKRPNNHTQ1320
1321 IVVYEINRTR TRHQKTCEFT IGYMAQHLQI LSDMRLVVAH QSGFTAYFLR GEATAMSLVH1380
1381 PENQLCAFLN YSGVDAVRVI EILCPSGGNF GEYLLVFQTL AIYVDLQGRK SRDREIMYPA1440
1441 FPTYITFCDG HLLVFSDTHL DIFNTQTAEW VQSIGLKQSL PLNNLGNVVL SSVNDTPLIV1500
1501 YLSNIHTKGL LQYRDGNRKG LPSIKRRFSI REINKTIKSD RRSKMISAPT NFNHISHMGP1560
1561 GDGIQNQRLL DLPTTLETAD QACSPIIHSL SCIPQSRKSN FLEQVDANSD DYGNDNIISR1620
1621 TPSPMASSFM DGLSNND |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
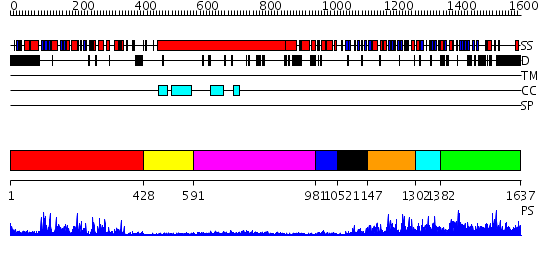
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..427] | 85.39794 | No description for 2f2uA was found. | |
| 2 | View Details | [428..590] | 2.2 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 3 | View Details | [591..980] | 11.522879 | Heavy meromyosin subfragment | |
| 4 | View Details | [981..1051] | 3.03 | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | |
| 5 | View Details | [1052..1146] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1147..1301] | 1.071968 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1302..1381] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1382..1637] | 1.05794 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)