













| General Information: |
|
| Name(s) found: |
retn-PA /
FBpp0071981
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 906 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQLRVHPTMD CSGRSTSNIE RDSDLGDDLS HGDRTDDEMR DCDSVDGEHH QLSAKAAIAA 60
61 RLSHTVSGGG GSFASPEPQT ELPLSHHHQL PPNHPLNALG SFMGIGGLHS IPNLQHSDVL 120
121 EKLKMQVRDM KVGLMEQDYA AAAHAAAFGA NMLPTTISSG FPLPHNSVAF GHVTSSPSGG 180
181 NGSSYNGGTT PTNSSNSNAT TNGGGTAGPG GTGGSGGGGG GGGGGGGGVG GHQFSFASPT 240
241 AAPSGKEANS ASNSSTSSEA SNSSQQNNGW SFEEQFKQVR QLYEINDDPK RKEFLDDLFS 300
301 FMQKRGTPIN RLPIMAKSVL DLYELYNLVI ARGGLVDVIN KKLWQEIIKG LHLPSSITSA 360
361 AFTLRTQYMK YLYPYECEKK NLSTPAELQA AIDGNRREGR RSSYGQYEAM HNQMPMTPIS 420
421 RPSLPGGMQQ MSPLALVTHA AVANNQQAQA AAAAAAAHHR LMGAPAFGQM PNLVKQEIES 480
481 RMMEYLQLIQ AKKEQGMPPV LGGNHPHQQQ HSQQQQQQQH HHQQQQQQQS QQQHHLQQQR 540
541 QRSQSPDLSK HEALSAQVAL WHMYHNNNSP PGSAHTSPQQ REALNLSDSP PNLTNIKRER 600
601 EREPTPEPVD QDDKFVDQPP PAKRVGSGLL PPGFPANFYL NPHNMAAVAA AAGFHHPSMG 660
661 HQQDAASEGE PEDDYAHGEH NTTGNSSSMH DDSEPQQMNG HHHHQTHHLD KSDDSAIENS 720
721 PTTSTTTGGS VGHRHSSPVS TKKKGGAKPQ SGGKDVPTED KDASSSGKLN PLETLSLLSG 780
781 MQFQVARNGT GDNGEPQLIV NLELNGVKYS GVLVANVPLS QSETRTSSPC HAEAPTVEEE 840
841 KDEEEEEEPK AAEEESHRSP VKQENEDVDQ DMEGSEVLLN GGASAVGGAG AGVGVGVPLL 900
901 KDAVVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
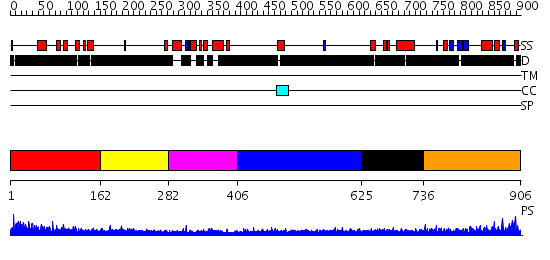
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [162..281] | 6.0 | Sec24 | |
| 3 | View Details | [282..405] | 18.0 | DNA-binding domain from the dead ringer protein | |
| 4 | View Details | [406..624] | 1.241996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [625..735] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [736..906] | 1.194995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)