













| General Information: |
|
| Name(s) found: |
comr-PA /
FBpp0071651
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 600 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
chromatin [NAS] |
| Biological Process: |
spermatogenesis
[IMP]
positive regulation of transcription from RNA polymerase II promoter [IMP] |
| Molecular Function: |
transcription activator activity
[IMP]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGNQDTLGQ SQQVNESGDE YEEDAATSWT NELEQQESRT PADSPEFLDF VDSEDPLEDF 60
61 ERNPLQKEPL DLAVKPVSDI NKKAAENHRR IYRQLGIHIP ETPKSHPLLH YAFVVKLFLA 120
121 TDNSSIHYLK WSANGRELEV DYQGMQEHLS SGCSMFHCRN TLQFTGTLLA HGFERVSLKD 180
181 VRKSSATNIT LIYRNPNFVK GQLLKLHKLA QECEKIPEIS SPGTAEPRRP PLHLPNYAHI 240
241 SRMVRRGDLC STPYTSRSAL QVARCHFQTL LGYQADLGVL KDRNNHDMFN KNIMTQLNVK 300
301 RGRSSINNAP VDLAPSKVSF EKFVKPHESV INIGIGQAPD YAGYYGQVEL FKVNEFFSEY 360
361 LPRYGSKITG YKEIVMDATN KSIGFQQNLP IGMNYSDDED DALPPISSDL DLDFMPSTSA 420
421 AAKMKSTGRK VVEDSDLEQA MQELCGGSNL NEEEELPQTS TRFKAKPKPR AKTKAKPTKR 480
481 QAKPVHSSSS ECEDFKPDEV YEGFKSRNPK NVKLKPSDEN ISYTLEEGTD FEEQVIEETT 540
541 ETKVQDSQYD DLDDDDDDEE EDYFDEDDED YVYKNLQYRN PPEPPKSRRY DLRNSKRNPR 600
601 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
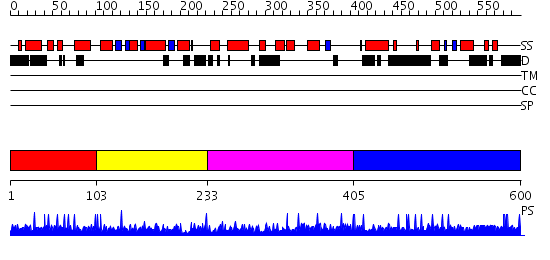
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [103..232] | 5.43 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 3 | View Details | [233..404] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [405..600] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)