













| General Information: |
|
| Name(s) found: |
CG6385-PA /
FBpp0088528
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 894 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
glycine catabolic process
[IEA]
|
| Molecular Function: |
sarcosine dehydrogenase activity
[ISS]
aminomethyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWRHRILQQA SRRGISKQVR EDGGARREPS GFLPGAADVV VIGGGSAGCH TLYHLARRGV 60
61 KAVLLERAQL TAGTTWHTAG LLWRLRPNDV DIQLLANSRR MLQQLEEETE LDPGWIQNGG 120
121 IFIAHNETRL DEYRRLATVG SALGIENQVL SPEDTQKLFP LLDPSAFVGA LYSPGDGVMD 180
181 PAMLCAALKK AATNLGAQVI ENCGVDDLLL EQTARGKKVV GVSTPFGDIK AEKVVNATGV 240
241 WGRDLVARHG THLPLVPMKH AYIVSESIPG VRGLPNIRDH DYSTYFRIQG DAICMGGYEP 300
301 NPILLEPVPK DFHFGLYELD WSVFETHVEG AQKLCPSYAK YGVKSTVCGP ESFTPDHKPL 360
361 MGPDPNLDGL YHNCGFNSAG MMFGGGCGEQ TALWVIQGQP DLPMFGFDLR RFTQEQGKAI 420
421 QWIREKSHES YVKNYSMVFK YDQPLAGRDF QKDPLHDEMM KAGAVMEEKQ GWERPGFFLP 480
481 TGSKKAVVQP YDWYGSYVHQ RHKDSEYERV LDGDLHYSRF SEYHDLIGSE ALACRNNAVV 540
541 FNMSYFAKLL LDGPQAQEAA DWLFSANTNR DPSKTVYTCA LNDAGGVEAD VTISRLAPGS 600
601 GEVYNPKING QGFYIVAGGA SAFYTYSVLL AEIRRKGFNA SLKDLTAELG VISIQGPNSR 660
661 KILQPLIDCD LSDEHVAPNS TRLAKFGDVG LRLLRVSFVG ELGYELHVPK KDCAAVYRSL 720
721 MKAGAGEDLR NAGYRSLYSL SSEKGYHLWS FDLRPDDTPL EAGLGFTCRK TGADYRGKAA 780
781 IENQRAEGLK KRLVYLTLRD QVPIWGLEGV YRNGEPVGIL RRAEYAYTLG KSLGQTYVSR 840
841 PDGKIIDADY IRNGEYEVDI LGKKYRADCH LRSPFDPTGQ RVLGNYASES KSNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
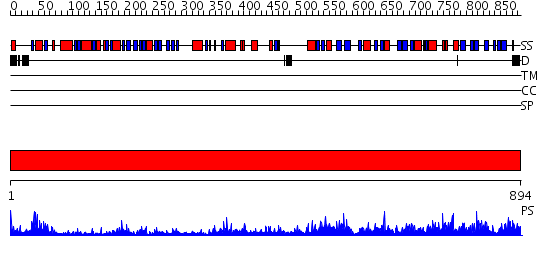
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..894] | 153.0 | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with acetate |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)