













| General Information: |
|
| Name(s) found: |
FBpp0309235
[FlyBase]
eIF3-S8-PA / FBpp0086013 [FlyBase] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 910 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[ISS]
eukaryotic translation initiation factor 3 complex [ISS] |
| Biological Process: |
translational initiation
[ISS]
|
| Molecular Function: |
translation initiation factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRFFANGSE SESESSEEEI QATNFNKASA FQFSDDEEEV KRVVRSTKEK RYENLTSIIK 60
61 TIRNHKKIKD IPNTLSSFED LTRAYQKALP VISKEENGIT PRFYIRCLAE LEDFINEVWE 120
121 DREGRKNLSK NNSKSLGTLR QKVRKYIKDF EDDLSRFREA PDQESEAEDE VVALESDGGD 180
181 AGDDSDAGVK PTEAVPKAVK SAPAKAAPAD DDDSDDSIDW DSDSESETES SDDENQYQNM 240
241 RERFLKRTTE KEEKDDDKRK DKRKEQKTKI RKRAEDDEDG EWETVVKGHV VEKPKMFEKD 300
301 AEIDVPLVLA KLLEIMSARG KKRTDRRLQI DLLFELRDIS DQHNLGTAVS VKIHFNIISA 360
361 IYDYNQKISE PMKLEHWALL LEVMQSMMKL LLANADIIMS ESVAEEHEEY ATSPFYVRGC 420
421 PLAAVERLDD EFVKLLKECD PHSNDYVSRL KDEVNVVKTI ELVLQYFERS GTNNERCRIY 480
481 LRKIEHLYYK FDPEVLKKKR GELPATTSTS VDVMDKLCKF IYAKDDTDRI RTRAILAHIY 540
541 HHAMHDNWFQ ARDLVLMSHL QDNIDAADPA TRILYNRMMA NLGLCAFRQG NVKDAHHCLV 600
601 DLMVTGKPKE LLAQGLLPQR QHERSAEQEK IEKQRQMPFH MHINLELLEC VYLVSAMLLE 660
661 IPYIAAHEFD ARRRMISKTF YQQLRSSERQ SLVGPPESMR EHVVAAAKAM RCGNWQACAN 720
721 FIVNKKMNTK VWDLFYESDR VREMLTKFIK EESLRTYLFT YSNVYTSISI PSLAQMYELP 780
781 VPKVHSIISK MIINEELMAS LDDPSETVGM HRSEPSRLQA LAMQFVDKVT NLVDVNEKVF 840
841 DMKQGNFFQR GNMGNRGDRG YNRNQNNQGG NWLGQRRDRN NRNRNQRGHH KNNQDRQQQQ 900
901 QQQVQTIDEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
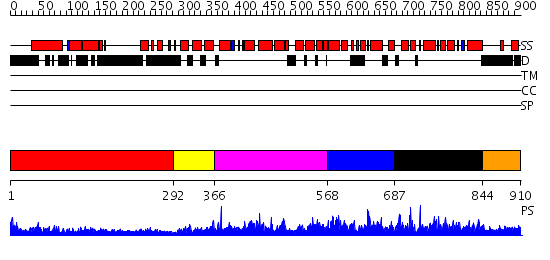
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..291] | 12.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [292..365] | 4.69897 | No description for 2i1jA was found. | |
| 3 | View Details | [366..567] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [568..686] | 1.051952 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [687..843] | 2.53 | Solution structure of the PCI domain | |
| 6 | View Details | [844..910] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)