













| General Information: |
|
| Name(s) found: |
RhoGEF2-PD /
FBpp0086125
[FlyBase]
RhoGEF2-PF / FBpp0086124 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2559 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lateral plasma membrane
[IDA]
apical part of cell [IDA] actomyosin contractile ring [IDA] basal part of cell [IDA] apical cortex [IDA] apical plasma membrane [IDA] actin cap [IDA] |
| Biological Process: |
regulation of cell shape
[IDA][IMP]
ventral furrow formation [IMP] regulation of Rho protein signal transduction [IEA] spiracle morphogenesis, open tracheal system [IMP] posterior midgut invagination [IMP] germ-band extension [IMP] actin filament organization [IMP] anterior midgut invagination [IMP] pole cell development [IMP] cellularization [IMP] gastrulation [TAS] regulation of axonogenesis [IGI] morphogenesis of embryonic epithelium [IMP] regulation of embryonic cell shape [IGI][IMP][NAS] actin cytoskeleton reorganization [IMP] pseudocleavage during syncytial blastoderm formation [IMP] |
| Molecular Function: |
protein binding
[IEA]
diacylglycerol binding [ISS] Rho guanyl-nucleotide exchange factor activity [IEA][ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDPSIKKRL LDLYTDEHEY DEVQEIPEES SIQPPETSTS HTSTNGSSHS GPGTATGPGA 60
61 TSAGPSAGAP QSPVIVVDSV PELPAPKQKS VKNSKSKQKQ KQLANKSKIP RSPSLASSLS 120
121 SLASSLSGHR DRDKDRDKDR ENQNAVPPQT PPLPPSYKQN QMNGDSTAAA GGGVSAPATP 180
181 TTANNNNASH NNGSIMGGGV QLNQSDNSNP VLQAPGERSS LNLTPLSRDL SGGHTQESTT 240
241 PATTPSTPSL ALPKNFQYLT LTVRKDSNGY GMKVSGDNPV FVESVKPGGA AEIAGLVAGD 300
301 MILRVNGHEV RLEKHPTVVG LIKASTTVEL AVKRSQKLTR PSSVSVVTPS TPILSGRDRT 360
361 ASITGPQPVD SIKRREMETY KIQTLQKMLE QEKLNLERLK SDQNNPSYKL SEANIRKLRE 420
421 QLHQVGAEDA PTVKLQAAAG NKNTALLTPN QIQHLSASAT HSNQQFHHLH HHHNLHNNNY 480
481 PPQQQPASTS PAFLSLLPRS LSSLSLGTRK NKTEKDLTTS SPFGLTTDFL QQQRMSHQAE 540
541 SMSQSMHQHT STPTSQQFFH PHQQQHRFKE TGPTSKGKNK FLISRSLIEE DVPPPLPQRN 600
601 PPRQLNLDLK NGNASPGGSH LVAPVSDLDR ATSPQLNRSQ QQQLPSSTDN SPSNAKSKRS 660
661 KIKTKALSDP KMSTQMFLQM ESASAAGAAG GSIEVDGGPP PLPPRLPGMM TEDMSRGSCQ 720
721 NLAQPNSVGT AFNYPLVSTT TAVQNDNLNI AFPLSQRPNI VQQLQQYQQQ QQHQMSGGQA 780
781 TGALGQTPNL GKNKHRRVGS SPDNMHPRHP DRITKTTSGS WEIVEKDGES SPPGTPPPPY 840
841 LSSSHMTVLE DPNENNRGAA AAGPGVFIES HQFTPMAGAS SPIPISLHSS HMHAAQSNDT 900
901 QKEIISMEDE NSDLDEPFID ENGPFNNLTR LLEAENVTFL AIFLNYVISN SDPAPLLFYL 960
961 ITELYKEGTS KDMRKWAYEI HSTFLVPRAP LSWYRQDESL AREVDNVLQL EYDKVEILRT1020
1021 VFLRSRKRAK DLISEQLREF QQKRTAGLGT IYGPTDDKLA EAKTDKLREQ IIDKYLMPNL1080
1081 HALIEDENGS PPEDVRKVAL CSALSTVIYR IFNTRPPPSS IVERVHHFVS RDKSFKSRIM1140
1141 GKNRKMNVRG HPLVLRQYYE VTHCNHCQTI IWGVSPQGYH CTDCKLNIHR QCSKVVDESC1200
1201 PGPLPQAKRL AHNDKISKFM GKIRPRTSDV IGNEKRSRQD EELDVELTPD RGQASIVRQP1260
1261 SDRRPDANIS IRSNGNTSCN TSGLNTTDLQ SSFHGSCAND SINPGGGAGC NMDLSTSVAS1320
1321 TTPSTSGSVA AGLSAFAELN ALDTVDKEAR RERYSQHPKH KSAPVSVNRS ESYKERLSNK1380
1381 RNRNSRRKTS DPSLSSRPND EQLDLGLSNA TYVGSSNSSL SSAGGSESPS TSMEHFAAPG1440
1441 AAGGVQVPPM GLNQNQHPHL LIQQHAQQYC QQDSFQAGLA GAAGSSAASN SSFWNAGHPL1500
1501 PVARWTLESE DEDDVNEADW SSMVAAEVLA ALTDAEKKRQ EIINEIYQTE RNHVRTLKLL1560
1561 DRLFFLPLYE SGLLSQDHLL LLFPPALLSL REIHGAFEQS LKQRRIEHNH VVNTIGDLLA1620
1621 DMFDGQSGVV LCEFAAQFCA RQQIALEALK EKRNKDEMLQ KLLKKSESHK ACRRLELKDL1680
1681 LPTVLQRLTK YPLLFENLYK VTVRLLPENT TEAEAIQRAV ESSKRILVEV NQAVKTAEDA1740
1741 HKLQNIQRKL DRSSYDKEEF KKLDLTQHHL IHDGNLTIKK NPSVQLHGLL FENMIVLLTK1800
1801 QDDKYYLKNL HTPLSITNKP VSPIMSIDAD TLIRQEAADK NSFFLIKMKT SQMLELRAPS1860
1861 SSECKTWFKH FSDVAARQSK NRSKNASSNH DTSISDPALA AIPHSNTKES LELSTDTVQP1920
1921 LAATATLTTT PLAPMLPIAT VTPAPATNNS NVSSLTGVQL RNPQRDATAS ESDADYVNTP1980
1981 KPRSSQNEVN RTMSIRSTGE PIQKYSANGT EANDVTLRHS QSTRESVRPG STGEERNSTY2040
2041 GMVGGNSKRD SASIVCSNNS NNTRTLLMQS PLVDPTAIQV SISPAHTAEP VLTPGEKLRR2100
2101 LDASIRNDLL EKQKIICDIF RLPVEHYDQI VDIAMMPEAP KDSADIALAA YDQIQTLTKM2160
2161 LNEYMHVTPE QEVSAVSTAV CGHCHEKEKL RKKVAPSSSF SSSPPPLPPP NRQHAQAQAQ2220
2221 IPPSRLMPKL QTLDLDEVAI HEDDDGYCEI DELRLPAIPS KPHERPTTPL APFNTEPKTS2280
2281 QSVIDASKRQ STDAVPEGLL EQEPLEGDKT ETKGEDNEVK TVPSDKLSES CNEERQCVEA2340
2341 DITKEVADPT TSKNEAAASV DELPSQSREI KTAENASKSV ADKKEDNEET IEEGVASTVD2400
2401 SSTQTSPTES PKETDKLTGG SSSTCGPNRI QHASVLEPSV PCHALSSIVT ILNEQISMLL2460
2461 PKINERDMER ERLRKENQHL RELLSALHDR QRVDEVKETP FDLKKLMHAE DVEFDDDIDA2520
2521 ISNSSLTPTP TPIPTASPSA SGQVETAEAM RITSTEDEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
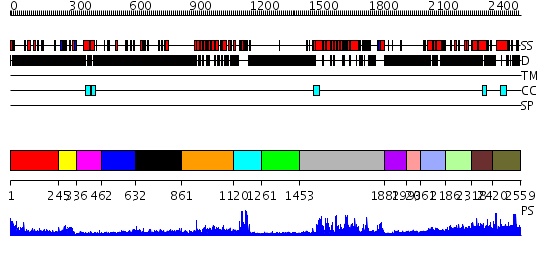
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..244] | 0.966 | Microvirus capsid proteins | |
| 2 | View Details | [245..335] | 15.39794 | No description for 2dlsA was found. | |
| 3 | View Details | [336..461] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [462..631] | 1.291993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [632..860] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [861..1119] | 35.09691 | Pdz-RhoGEF RGS-like domain | |
| 7 | View Details | [1120..1260] | 23.0 | Crystal Structure of the Human Beta2-Chimaerin | |
| 8 | View Details | [1261..1452] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1453..1881] | 82.522879 | Crystal Structure of the DH/PH domains of Leukemia-associated RhoGEF | |
| 10 | View Details | [1882..1992] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1993..2060] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [2061..2185] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [2186..2317] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [2318..2419] | 1.023993 | View MSA. No confident structure predictions are available. | |
| 15 | View Details | [2420..2559] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 14 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 15 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)