













| General Information: |
|
| Name(s) found: |
RhoGEF2-PE /
FBpp0086123
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 2205 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lateral plasma membrane
[IDA]
apical part of cell [IDA] actomyosin contractile ring [IDA] basal part of cell [IDA] apical cortex [IDA] apical plasma membrane [IDA] actin cap [IDA] |
| Biological Process: |
regulation of cell shape
[IDA][IMP]
ventral furrow formation [IMP] regulation of Rho protein signal transduction [IEA] spiracle morphogenesis, open tracheal system [IMP] posterior midgut invagination [IMP] germ-band extension [IMP] actin filament organization [IMP] anterior midgut invagination [IMP] pole cell development [IMP] cellularization [IMP] gastrulation [TAS] regulation of axonogenesis [IGI] morphogenesis of embryonic epithelium [IMP] regulation of embryonic cell shape [IGI][IMP][NAS] actin cytoskeleton reorganization [IMP] pseudocleavage during syncytial blastoderm formation [IMP] |
| Molecular Function: |
protein binding
[IEA]
diacylglycerol binding [ISS] Rho guanyl-nucleotide exchange factor activity [IEA][ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDPSIKKRL LDLYTDEHEY DEVQEIPEES SIQPPETSTS HTSTNGSSHS GPGTATGPGA 60
61 TSAGPSAGAP QSPVIVVDSV PELPAPKQKS VKNSKSKQKQ KQLANKSKIP RSPSLASSLS 120
121 SLASSLSGHR DRDKDRDKDR ENQNAVPPQT PPLPPSYKQN QMNGDSTAAA GGGVSAPATP 180
181 TTANNNNASH NNGSIMGGGV QLNQSDNSNP VLQAPGERSS LNLTPLSRDL SGGHTQESTT 240
241 PATTPSTPSL ALPKNFQYLT LTVRKDSNGY GMKVSGDNPV FVESVKPGGA AEIAGLVAGD 300
301 MILRVNGHEV RLEKHPTVVG LIKASTTVEL AVKRSQKLTR PSSVSVVTPS TPILSGRDRT 360
361 ASITGPQPVD SIKRREMETY KIQTLQKMLE QEKLNLERLK SDQNNPSYKL SEANIRKLRE 420
421 QLHQVGAEAL GQTPNLGKNK HRRVGSSPDN MHPRHPDRIT KTTSGSWEIV EKDGESSPPG 480
481 TPPPPYLSSS HMTVLEDPNE NNRGAAAAGP GVFIESHQFT PMAGASSPIP ISLHSSHMHA 540
541 AQSNDTQKEI ISMEDENSDL DEPFIDENGP FNNLTRLLEA ENVTFLAIFL NYVISNSDPA 600
601 PLLFYLITEL YKEGTSKDMR KWAYEIHSTF LVPRAPLSWY RQDESLAREV DNVLQLEYDK 660
661 VEILRTVFLR SRKRAKDLIS EQLREFQQKR TAGLGTIYGP TDDKLAEAKT DKLREQIIDK 720
721 YLMPNLHALI EDENGSPPED VRKVALCSAL STVIYRIFNT RPPPSSIVER VHHFVSRDKS 780
781 FKSRIMGKNR KMNVRGHPLV LRQYYEVTHC NHCQTIIWGV SPQGYHCTDC KLNIHRQCSK 840
841 VVDESCPGPL PQAKRLAHND KISKFMGKIR PRTSDVIGNE KRSRQDEELD VELTPDRGQA 900
901 SIVRQPSDRR PDANISIRSN GNTSCNTSGL NTTDLQSSFH GSCANDSINP GGGAGCNMDL 960
961 STSVASTTPS TSGSVAAGLS AFAELNALDT VDKEARRERY SQHPKHKSAP VSVNRSESYK1020
1021 ERLSNKRNRN SRRKTSDPSL SSRPNDEQLD LGLSNATYVG SSNSSLSSAG GSESPSTSME1080
1081 HFAAPGAAGG VQVPPMGLNQ NQHPHLLIQQ HAQQYCQQDS FQAGLAGAAG SSAASNSSFW1140
1141 NAGHPLPVAR WTLESEDEDD VNEADWSSMV AAEVLAALTD AEKKRQEIIN EIYQTERNHV1200
1201 RTLKLLDRLF FLPLYESGLL SQDHLLLLFP PALLSLREIH GAFEQSLKQR RIEHNHVVNT1260
1261 IGDLLADMFD GQSGVVLCEF AAQFCARQQI ALEALKEKRN KDEMLQKLLK KSESHKACRR1320
1321 LELKDLLPTV LQRLTKYPLL FENLYKVTVR LLPENTTEAE AIQRAVESSK RILVEVNQAV1380
1381 KTAEDAHKLQ NIQRKLDRSS YDKEEFKKLD LTQHHLIHDG NLTIKKNPSV QLHGLLFENM1440
1441 IVLLTKQDDK YYLKNLHTPL SITNKPVSPI MSIDADTLIR QEAADKNSFF LIKMKTSQML1500
1501 ELRAPSSSEC KTWFKHFSDV AARQSKNRSK NASSNHDTSI SDPALAAIPH SNTKESLELS1560
1561 TDTVQPLAAT ATLTTTPLAP MLPIATVTPA PATNNSNVSS LTGVQLRNPQ RDATASESDA1620
1621 DYVNTPKPRS SQNEVNRTMS IRSTGEPIQK YSANGTEAND VTLRHSQSTR ESVRPGSTGE1680
1681 ERNSTYGMVG GNSKRDSASI VCSNNSNNTR TLLMQSPLVD PTAIQVSISP AHTAEPVLTP1740
1741 GEKLRRLDAS IRNDLLEKQK IICDIFRLPV EHYDQIVDIA MMPEAPKDSA DIALAAYDQI1800
1801 QTLTKMLNEY MHVTPEQEVS AVSTAVCGHC HEKEKLRKKV APSSSFSSSP PPLPPPNRQH1860
1861 AQAQAQIPPS RLMPKLQTLD LDEVAIHEDD DGYCEIDELR LPAIPSKPHE RPTTPLAPFN1920
1921 TEPKTSQSVI DASKRQSTDA VPEGLLEQEP LEGDKTETKG EDNEVKTVPS DKLSESCNEE1980
1981 RQCVEADITK EVADPTTSKN EAAASVDELP SQSREIKTAE NASKSVADKK EDNEETIEEG2040
2041 VASTVDSSTQ TSPTESPKET DKLTGGSSST CGPNRIQHAS VLEPSVPCHA LSSIVTILNE2100
2101 QISMLLPKIN ERDMERERLR KENQHLRELL SALHDRQRVD EVKETPFDLK KLMHAEDVEF2160
2161 DDDIDAISNS SLTPTPTPIP TASPSASGQV ETAEAMRITS TEDEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
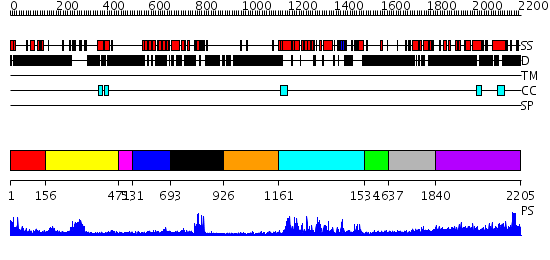
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | 1.251994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [156..470] | 22.0 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 3 | View Details | [471..530] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [531..692] | 5.62 | Pdz-RhoGEF RGS-like domain | |
| 5 | View Details | [693..925] | 20.0 | Crystal Structure of the Human Beta2-Chimaerin | |
| 6 | View Details | [926..1160] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1161..1533] | 65.09691 | Crystal Structure of the DH/PH domains of Leukemia-associated RhoGEF | |
| 8 | View Details | [1534..1636] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1637..1839] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1840..2205] | 7.154902 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)