













| General Information: |
|
| Name(s) found: |
FBpp0308554
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 897 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[ISS]
|
| Biological Process: |
peroxisome organization
[ISS]
|
| Molecular Function: |
ATP binding
[IEA]
nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNENDDAKKV VGKRNSVSFD QQDPGQPRFS FSGRVQELVE GYNSAIQLLA PKWPWFLYPG 60
61 YLTYAVYWRY AKYYKGKFLL TPIPNRLYDL LIKNEQIKSE HLVFASQEVY ENYAESPDVN 120
121 FVNLVIRGRR VRIGKPLASD EGNMTQTVIK QMLQEAPHTI VAQILPAAQC QPNCLYVQEN 180
181 FYGNILDRFQ IRSNCWVQVE HFADEQTIPG IATKAHVYLV ADPHELPAEL TEVILSNYFN 240
241 TPRLMHRGHT YRIEVNAELV GTATYAHYYL IFAHLRHVHL KCIHLETKGS EFELQAIVAK 300
301 NFSNLVQVPA SHSFLPRQVL DSMAIVENYP SGLRKPYKLL RSSVDAFLPK KSACLSSKHI 360
361 FPVFLLQGER GSGKSKLVSA VAQELGMHIY GADCAEIVSQ VPSHTEMKLK AVFAKSQVCE 420
421 PLMICFHNFE IFGIDNEGNE DLRLLSAFHV QVQELFNRDR KHPIVVVALT SDRHLKPMIQ 480
481 GLFLEIINID MPSKEERFEI LRWMHVRETF NDIIYNQKAI EQLPLFSREN QSQFMPRISP 540
541 SWRETLNVLQ DVAAKSQGFL LGDLQLLYDN AVRMKIRNRL GRTTLDMSHF AKNLTDMQSS 600
601 FADSLGAPKV PKVYWSDIGG LAKLKDEIQS SIGLPLKHVH LMGKNLRRSG ILLYGPPGTG 660
661 KTLVAKAVAT ECNLSFLSVQ GPELLNMYVG QSEQNVREVF SRARSAAPCV LFLDELDSLA 720
721 PNRGVAGDSG GVMDRVVSQL LAEMDGMSDG DTSKPIFILA ATNRPDLIDP ALLRPGRFDK 780
781 LFYVGPCSTA EDKAAVLRAQ TQRFALDAGV DMEQIAERLK SEMSGADLYS ICSNAWLSAV 840
841 RRTIDGHLSG TISEKELVPE NVIVQEEDFT KSFNKFVPSI SAKDLEYFNN LKASYSV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
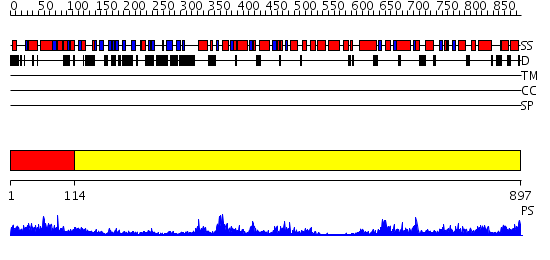
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 5.154902 | Crystal structure of N-terminal domain of E.Coli Lon Protease | |
| 2 | View Details | [114..897] | 156.0 | VCP/p97 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)