













| General Information: |
|
| Name(s) found: |
sxc-PC /
FBpp0085422
[FlyBase]
sxc-PA / FBpp0085421 [FlyBase] sxc-PB / FBpp0085420 [FlyBase] |
| Description(s) found:
Found 51 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1059 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
wing disc development
[IMP]
protein amino acid glycosylation [IMP] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[NAS]
acetylglucosaminyltransferase activity [IMP] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHVEQTRINM QSQGQSHQLP SAAHILLDQN PNSTGSNLVV KQNDIQSLSS VGLLELAHRE 60
61 YQAVDYESAE KHCMQLWRQD STNTGVLLLL SSIHFQCRRL DKSAQFSTLA IKQNPVLAEA 120
121 YSNLGNVFKE RGQLQEALDN YRRAVRLKPD FIDGYINLAA ALVAARDMES AVQAYITALQ 180
181 YNPDLYCVRS DLGNLLKALG RLEEAKACYL KAIETCPGFA VAWSNLGCVF NAQGEIWLAI 240
241 HHFEKAVTLD PNFLDAYINL GNVLKEARIF DRAVAAYLRA LNLSPNNAVV HGNLACVYYE 300
301 QGLIDLAIDT YRRAIELQPN FPDAYCNLAN ALKEKGQVKE AEDCYNTALR LCSNHADSLN 360
361 NLANIKREQG YIEEATRLYL KALEVFPDFA AAHSNLASVL QQQGKLKEAL MHYKEAIRIQ 420
421 PTFADAYSNM GNTLKELQDV SGALQCYTRA IQINPAFADA HSNLASIHKD SGNIPEAIQS 480
481 YRTALKLKPD FPDAYCNLAH CLQIVCDWTD YDIRMKKLVS IVTEQLEKNR LPSVHPHHSM 540
541 LYPLTHDCRK AIAARHANLC LEKVHVLHKK PYNFLKKLPT KGRLRIGYLS SDFGNHPTSH 600
601 LMQSVPGLHD RSKVEIFCYA LSPDDGTTFR HKISRESENF VDLSQIPCNG KAADKIFNDG 660
661 IHILVNMNGY TKGARNEIFA LRPAPIQVMW LGYPGTSGAS FMDYIITDSV TSPLELAYQY 720
721 SEKLSYMPHT YFIGDHKQMF PHLKERIIVC DKQQSSVVDN VTVINATDLS PLVENTDVKE 780
781 IKEVVNAQKP VEITHKVAEL PNTTQIVSMI ATGQVQTSLN GVVVQNGLAT TQTNNKAATG 840
841 EEVPQNIVIT TRRQYMLPDD AVVYCNFNQL YKIDPQTLES WVEILKNVPK SVLWLLRFPA 900
901 VGEQNIKKTV SDFGISPDRV IFSNVAAKEE HVRRGQLADI CLDTPLCNGH TTSMDVLWTG 960
961 TPVVTLPGET LASRVAASQL ATLGCPELIA RTREEYQNIA IRLGTKKEYL KALRAKVWKA1020
1021 RVESPLFDCS QYAKGLEKLF LRMWEKYENG ELPDHISAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
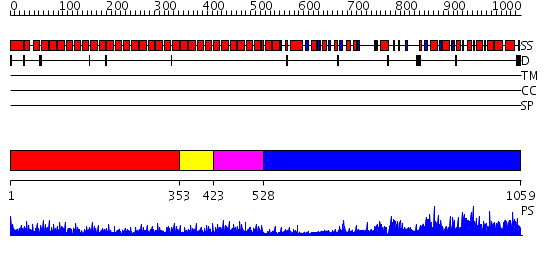
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..352] | 80.522879 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | |
| 2 | View Details | [353..422] | 4.6 | Hop | |
| 3 | View Details | [423..527] | 34.09691 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 4 | View Details | [528..1059] | 2.67 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)