













| General Information: |
|
| Name(s) found: |
gi|157928304
[NCBI NR]
gi|123996267 [NCBI NR] gi|123981428 [NCBI NR] |
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 346 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METQLSNGPT CNNTANGPTT INNNCSSPVD SGNTEDSKTN LIVNYLPQNM TQEELKSLFG 60
61 SIGEIESCKL VRDKITGQSL GYGFVNYIDP KDAEKAINTL NGLRLQTKTI KVSYARPSSA 120
121 SIRDANLYVS GLPKTMTQKE LEQLFSQYGR IITSRILVDQ VTGISRGVGF IRFDKRIEAE 180
181 EAIKGLNGQK PPGATEPITV KFANNPSQKT NQAILSQLYQ SPNRRYPGPL AQQAQRFRFS 240
241 PMTIDGMTSL AGINIPGHPG TGWCIFVYNL APDADESILW QMFGPFGAVT NVKVIRDFNT 300
301 NKCKGFGFVT MTNYDEAAMA IASLNGYRLG DRVLQVSFKT NKTHKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
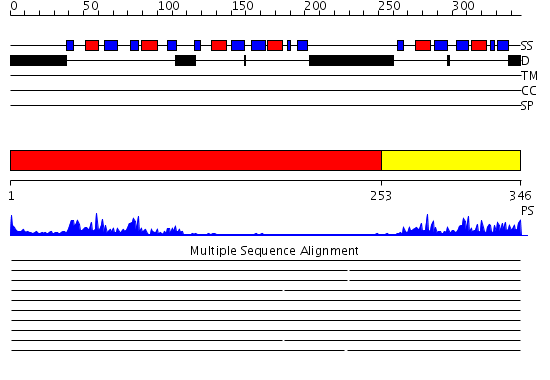
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..252] | 20.09691 | Hu antigen C (Huc) | |
| 2 | View Details | [253..346] | 19.39794 | Hu antigen C (Huc) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)