













| General Information: |
|
| Name(s) found: |
gi|123997931
[NCBI NR]
gi|123983224 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 525 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLFTANPFE QDVEKATNEY NTTEDWSLIM DICDKVGSTP NGAKDCLKAI MKRVNHKVPH 60
61 VALQALTLLG ACVANCGKIF HLEVCSRDFA TEVRAVIKNK AHPKVCEKLK SLMVEWSEEF 120
121 QKDPQFSLIS ATIKSMKEEG ITFPPAGSQT VSAAAKNGTS SNKNKEDEDI AKAIELSLQE 180
181 QKQQHTETKS LYPSSEIQLN NKVARKVRAL YDFEAVEDNE LTFKHGEIII VLDDSDANWW 240
241 KGENHRGIGL FPSNFVTTNL NIETEAAAVD KLNVIDDDVE EIKKSEPEPV YIDEDKMDRA 300
301 LQVLQSIDPT DSKPDSQDLL DLEDICQQMG PMIDEKLEEI DRKHSELSEL NVKVLEALEL 360
361 YNKLVNEAPV YSVYSKLHPP AHYPPASSGV PMQTYPVQSH GGNYMGQSIH QVTVAQSYSL 420
421 GPDQIGPLRS LPPNVNSSVT AQPAQTSYLS TGQDTVSNPT YMNQNSNLQS ATGTTAYTQQ 480
481 MGMSVDMSSY QNTTSNLPQL AGFPVTVPAH PVAQQHTNYH QQPLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
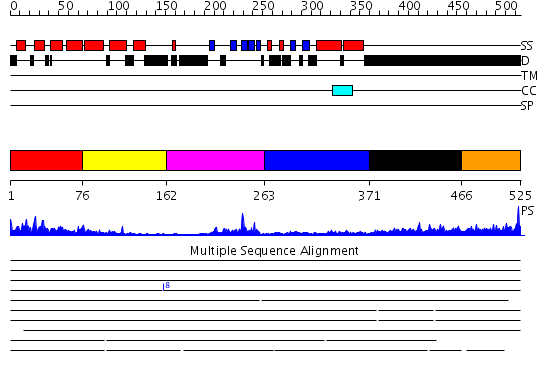
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | 40.69897 | The solution structure of the VHS domain of human Signal transducing adaptor molecule 2 | |
| 2 | View Details | [76..161] | 3.67 | The solution structure of the VHS domain of human Signal transducing adaptor molecule 2 | |
| 3 | View Details | [162..262] | 12.09691 | Solution structure of the SH3 domain of the Signal transducing adaptor molecule 2 | |
| 4 | View Details | [263..370] | 10.522879 | NMR ENSEMBLE OF BLK SH2 DOMAIN, 20 STRUCTURES | |
| 5 | View Details | [371..465] | 1.06 | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | |
| 6 | View Details | [466..525] | 3.161995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)