













| General Information: |
|
| Name(s) found: |
gi|123998539
[NCBI NR]
gi|123993693 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRRTRCEDL DELHYQDTDS DVPEQRDSKC KVKWTHEEDE QLRALVRQFG QQDWKFLASH 60
61 FPNRTDQQCQ YRWLRVLNPD LVKGPWTKEE DQKVIELVKK YGTKQWTLIA KHLKGRLGKQ 120
121 CRERWHNHLN PEVKKSCWTE EEDRIICEAH KVLGNRWAEI AKMLPGRTDN AVKNHWNSTI 180
181 KRKVDTGGFL SESKDCKPPV YLLLELEDKD GLQSAQPTEG QGSLLTNWPS VPPTIKEEEN 240
241 SEEELAAATT SKEQEPIGTD LDAVRTPEPL EEFPKREDQE GSPPETSLPY KWVVEAANLL 300
301 IPAVGSSLSE ALDLIESDPD AWCDLSKFDL PEEPSAEDSI NNSLVQLQAS HQQQVLPPRQ 360
361 PSALVPSVTE YRLDGHTISD LSRSSRGELI PISPSTEVGG SGIGTPPSVL KRQRKRRVAL 420
421 SPVTENSTSL SFLDSCNSLT PKSTPVKTLP FSPSQFLNFW NKQDTLELES PSLTSTPVCS 480
481 QKVVVTTPLH RDKTPLHQKH AAFVTPDQKY SMDNTPHTPT PFKNALEKYG PLKPLPQTPH 540
541 LEEDLKEVLR SEAGIELIIE DDIRPEKQKR KPGLRRSPIK KVRKSLALDI VDEDVKLMMS 600
601 TLPKSLSLPT TAPSNSSSLT LSGIKEDNSL LNQGFLQAKP EKAAVAQKPR SHFTTPAPMS 660
661 SAWKTVACGG TRDQLFMQEK ARQLLGRLKP SHTSRTLILS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
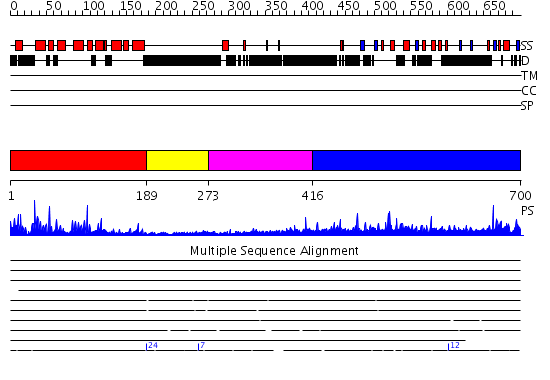
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..188] | 49.154902 | c-Myb, DNA-binding domain | |
| 2 | View Details | [189..272] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [273..415] | 1.302993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [416..700] | 104.39794 | No description for PF09316.1 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)