













| General Information: |
|
| Name(s) found: |
gi|168277836
[NCBI NR]
gi|123995817 [NCBI NR] gi|123981010 [NCBI NR] |
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 548 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATSSEEVLL IVKKVRQKKQ DGALYLMAER IAWAPEGKDR FTISHMYADI KCQKISPEGK 60
61 AKIQLQLVLH AGDTTNFHFS NESTAVKERD AVKDLLQQLL PKFKRKANKE LEEKNRMLQE 120
121 DPVLFQLYKD LVVSQVISAE EFWANRLNVN ATDSSSTSNH KQDVGISAAF LADVRPQTDG 180
181 CNGLRYNLTS DIIESIFRTY PAVKMKYAEN VPHNMTEKEF WTRFFQSHYF HRDRLNTGSK 240
241 DLFAECAKID EKGLKTMVSL GVKNPLLDLT ALEDKPLDEG YGISSVPSAS NSKSIKENSN 300
301 AAIIKRFNHH SAMVLAAGLR KQEAQNEQTS EPSNMDGNSG DADCFQPAVK RAKLQESIEY 360
361 EDLGKNNSVK TIALNLKKSD RYYHGPTPIQ SLQYATSQDI INSFQSIRQE MEAYTPKLTQ 420
421 VLSSSAASST ITALSPGGAL MQGGTQQAIN QMVPNDIQSE LKHLYVAVGE LLRHFWSCFP 480
481 VNTPFLEEKV VKMKSNLERF QVTKLCPFQE KIRRQYLSTN LVSHIEEMLQ TAYNKLHTWQ 540
541 SRRLMKKT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
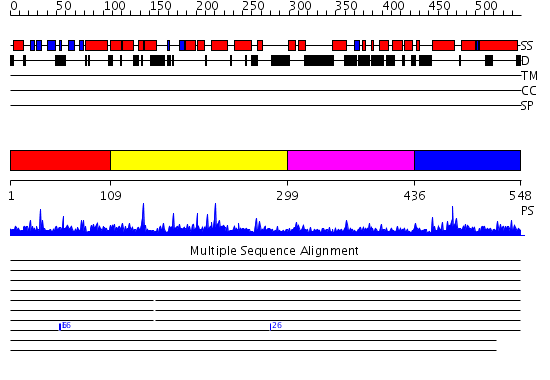
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 46.0 | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | |
| 2 | View Details | [109..298] | 18.008774 | No description for PF03909.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [299..435] | 1.036979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [436..548] | 2.03499 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)