













| General Information: |
|
| Name(s) found: |
YH7G_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSGDVSRRQ RVSRACDECH RRKIKCDQRR PCSNCIAYNY ECTYGQPFKR LRHAPEKYIE 60
61 FLELRLKYLR GLAEESDPNL KLPSFLAPPN DKDSPVNQSP WKRSDSSKRS SSQDEFESLF 120
121 DRYGQLSLKD DGKADFRGSS SGFVFMKNIH QNIARNSTVP NPVQESNSSS SQPDPLSFPY 180
181 LPPTPAEDEH KKPPLKIQLP PYEEALSIVS QFFMNDHFLV HIHHPASFFE KMHMYYKTGK 240
241 TDNNFHFLLV ATLCLGYTYM PDESPSANYP YHEAYEYYYY IRSSFSWEDS YTIEVVQILL 300
301 SVALFALFSS RLSQAYTFTN NALLCCHELG LHKDFSDVLT SHESRLSKRV FYSVYVLACY 360
361 TSTIVGLPLS IEDVDIDQSL PNSFDFTLEN DQVPPRLIAS ECTSLEVFIQ HITLSRILSH 420
421 FVRKVYPVKS PSDSHCKVSL PAVRDHEEKL TYWWKNLPSY LKMSEVPKFS PKWIQAIILE 480
481 LKFRQIELIF YRPFIHSIST PIDNQNGSPM KPANFALKCA QSAERVVFLL QELAKSPNTP 540
541 KLFFNLYSGY YALMTLTYCA TLTKDDANKS NNFITKARLG FHCLQMIYRE STYYSTIMEA 600
601 IKNLLIAYDM NSSGTENLDA TPDVTGQLPN NFSQRTSNIP REFPQAQIFY SDAPYPGYYN 660
661 PAQFQNAPTN FPMPTYGGRT QDQSYPRQNG YPSYSDGNVY PHDRVMINYG SSMPTANGFY 720
721 VPNTYSPVPF PYNTSYPPYM SPTSNMPQAF QAYSQYPYQH PPFPLSEQML PLPTSGVMMA 780
781 PGAAKSGMPY PFIQPPSMTN QVAYPTVRDG SNNSPDHPSS SNSKRTE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
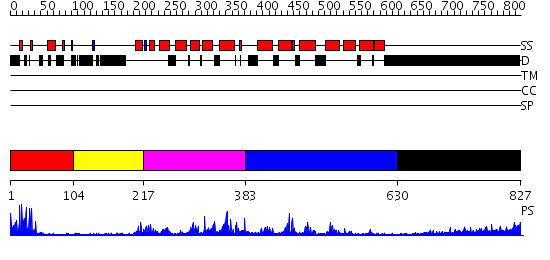
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | 7.39794 | PPR1 | |
| 2 | View Details | [104..216] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [217..382] | 24.853872 | No description for PF04082.9 was found. Confident ab initio structure predictions are available. | |
| 4 | View Details | [383..629] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [630..827] | 12.818997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.52 |
Source: Reynolds et al. (2008)