













| General Information: |
|
| Name(s) found: |
YCN3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 867 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLNTLPFHS INSVNVSSAP TVNERSYPFL LDASAGVSGF NPVVSSPEKK ARMKYKKNST 60
61 SPNMDVKSRK KVSRACDFCR QKKIRCDMDQ SPRPGNACIN CRKHHLDCNF TRTPLKRGPA 120
121 KGFNRNADEK QKRASGSAKS SSPAVNGSVF SGNEASPSSR APSITPVDSV NTTTSAIQVP 180
181 SVTLTAPAPL GVDQKISQDQ KPDSWLTYNA QFAQNSPQLA PSIPSPMKLS PANQQAMPPY 240
241 PQMLGPGSIS SYTNSNLGPS AGFRPPTFFS SPSPQPYSGP ILASTAPTLD GSYLSNPSNS 300
301 NPAVMSLSSN FPSPPKPNNP VYLPPRGNPT VNDRVSNVLP SITSFDSSVT TVPSNSPATL 360
361 NSYTTSVPSG MSRHPMLMNP STPEPSLGVN SPSLRPLQSL NNVQNSYRVA STQAPPPHPL 420
421 RNYTSDAESI SMRSKSTQAS DAATFREVEQ LYQENVEWDD AAIDRYYLLI HSTLPILHHS 480
481 KARLKSELEK APINLRSSCL HAIYSLVNRP PFATLGHVFH NTPMKAIGLL NLICSNVQDL 540
541 SNRILHLQTM ILLAIESDQR GPTTITGRNG LPQGMWLGAA IGLACNMRLH IQSHLSLQSI 600
601 NEDMDSDEAL CRRAWWVLVV LDRWHSMSTC SPLFLPETFI NLTIQDQKLL GTFPSQLVRL 660
661 SLIVGHISDV FQSPDPTDRQ SPIVTQQLRS EIDAFRQSVD VVWGQMNLLT LAVTHVKVLL 720
721 ELCINARPST VLVPAMKMAT ILSSSSTPMT PLNHHFFSLA TCVLIGVFDL PELQNEARRG 780
781 LEHIRECIEK RRDIVSREDH EDWDYIVLKL INAKMQGMPI NSDPSIPPHV PPSSAFAYSN 840
841 QEMDSATFKD AYLYTRLCNL GYLGFLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
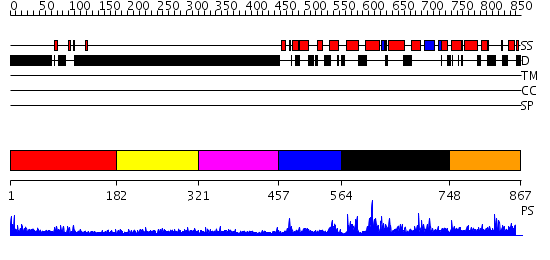
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | 2.34 | PPR1 | |
| 2 | View Details | [182..320] | 1.317993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [321..456] | 4.335994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [457..563] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [564..747] | 1.131993 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [748..867] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)