













| General Information: |
|
| Name(s) found: |
BZZ1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLETYKFSD ELHDDFKVVD SWINNGAKWL EDIQLYYKER SSIEKEYAQK LASLSNKYGE 60
61 KKSRKSSALS VGDTPAMSAG SLECASLTTW SKILDELTRS SKTHQKLSDD YSLDIAEKLK 120
121 KLESHIEALR KVYDDLYKKF SSEKETLLNS VKRAKVSYHE ACDDLESARQ KNDKYREQKT 180
181 QRNLKLSESD MLDKKNKYLL RMLVYNAHKQ KFYNETLPTL LNHMQVLNEY RVSNLNEIWC 240
241 NSFSIEKSLH DTLSQRTVEI QSEIAKNEPV LDSAMFGRHN SKNWALPADL HFEPSPIWHD 300
301 TDALVVDGSC KNYLRNLLVH SKNDLGKQKG ELVSLDSQLE GLRVDDPNSA NQSFESKKAS 360
361 INLEGKELMV KARIEDLEVR INKITSVANN LEEGGRFHDF KHVSFKLPTS CSYCREIIWG 420
421 LSKRGCVCKN CGFKCHARCE LLVPANCKNG EPEVADDDAV DTSVTATDDF DASASSSNAY 480
481 ESYRNTYTDD MDSSSIYQTS LSNVKTEETT PAEPASKVDG VVLYDFTGEH EGVITASEGQ 540
541 EFTLLEPDDG SGWVRVKIDG TDGLIPASYV KLNDELNTSV TLDGDSSYVK ALYAYTAQSD 600
601 MELSIQEGDI IQVTNRNAGN GWSEGILNGV TGQFPANYVT DV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
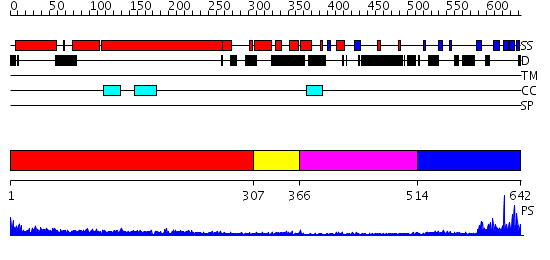
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..306] | 16.522879 | No description for 2eflA was found. | |
| 2 | View Details | [307..365] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [366..513] | 2.85 | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | |
| 4 | View Details | [514..642] | 15.69897 | Growth factor receptor-bound protein 2 (GRB2), N- and C-terminal domains; Growth factor receptor-bound protein 2 (GRB2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)