













| General Information: |
|
| Name(s) found: |
YAD7_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 527 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYPYCLKYLT NGLRSKQMIN FLCIQNILSA RTLKICRFRF LPTPSHPTHK MAQETRAISS 60
61 ISEAANKRAR KLSKRRAKKP VEEGSSGHVL LLEIENLIEK PVLANVPFER FQEIEVKISY 120
121 LSSSGDGIGL VCNDQYAVVV PFTLPGDIVK AKLHFLADTY ALADFLEVIS PSEDRDDTLI 180
181 KCPYFAKCGG CQYQMLSYDK QLLQKKRVVE KAFQYYSKLD SSLLPAIKDT VGSPLQYNYR 240
241 TKITPHFDVP KGGTKGPLII GFQEKGRRRV MDIEECPIAT KTINEEYPKI IEDVQSRANT 300
301 FKRGATILMR DSATEDGKHC VITDHKMIVR EQFGDLSFTF PAGAFFQNNN SILEKFTSYV 360
361 REQLLNPFGR KEHQRPKYFV DAYCGSGLFS VACSKGFLSV IGVEISADSV RYAEENAKRN 420
421 NVSNATFIVG QAEKIFSSIE TPPNETAMVI DPPRKGCDQS FLNQLLEYSP YRIVYISCNV 480
481 HTQARDVGFL LSQEKGRSYK IDEIRGFDLF PQSHHVESIL TLTKVVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
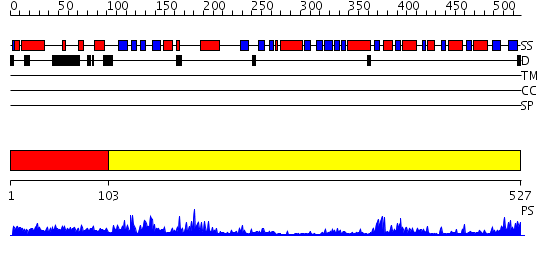
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 6.39794 | Structure of archaeal translation factor aIF2beta from Methanobacterium thermoautrophicum | |
| 2 | View Details | [103..527] | 66.39794 | Crystal Structure of RumA, the iron-sulfur cluster containing E. coli 23S Ribosomal RNA 5-Methyluridine Methyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)