













| General Information: |
|
| Name(s) found: |
NKX31_MOUSE
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Mus musculus |
| Length: | 237 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
negative regulation of epithelial cell proliferation involved in prostate gland development
[IMP]
branching involved in prostate gland morphogenesis [IMP] transcription [IEA] internal genitalia morphogenesis [TAS] multicellular organismal development [IEA] negative regulation of cell proliferation [IGI] regulation of transcription, DNA-dependent [IDA] salivary gland development [IMP] branching morphogenesis of a tube [IMP] regulation of transcription [IEA] epithelial cell proliferation involved in salivary gland morphogenesis [IMP] pharyngeal system development [IEP negative regulation of insulin-like growth factor receptor signaling pathway [IDA] heart development [IEP urogenital system development [TAS] somitogenesis [IEP negative regulation of epithelial cell proliferation [IMP] prostate gland development [TAS] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [IDA] protein binding [IPI] transcription regulator activity [IEA] sequence-specific DNA binding [IEA] transcription factor binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRVAEPREP RVEAGGRSPW AAPPTQSKRL TSFLIQDILR DRAERHGGHS GNPQHSPDPR 60
61 RDSAPEPDKA GGRGVAPEDP PSIRHSPAET PTEPESDAHF ETYLLDCEHN PGDLASAPQV 120
121 TKQPQKRSRA AFSHTQVIEL ERKFSHQKYL SAPERAHLAK NLKLTETQVK IWFQNRRYKT 180
181 KRKQLSEDLG VLEKNSPLSL PALKDDSLPS TSLVSVYTSY PYYPYLYCLG SWHPSFW |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
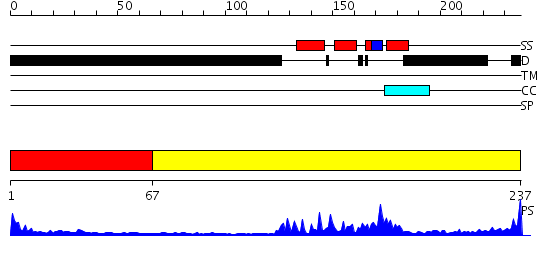
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [67..237] | 22.522879 | Homeobox protein hox-b1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)