













| General Information: |
|
| Name(s) found: |
DPO5_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 959 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATKTQLELF TKLTSNDKAI RLSSAAQLID SLSNEEELKY SLNRLTKGLS SGRESARIGF 60
61 AVALTELLTR TKDIRATHVL DLLVKHNTAS GNLKGQDERD FYFGLLFGLQ SIVYSGILTH 120
121 KESTIEDFQR VVDLLLQLSG KKNWLQDVCF YVIYKLVEQI PEISFSSNAF LAVNKLLQTP 180
181 AVSKSTEGVG LFLCLTRVPD NVKSEEVAMA NWEPAHPLHK SNLVTLSKIM RQADASETGG 240
241 QNSAWKQKIP MVWKYIFEEY QRKTYSGLAP FHDFWAVVVD EGIFSSTSSL ERKFWGFQIM 300
301 ELALDYVSSD NIGDIFSKNF LHCLINHLSD EDRYLYRAAK RVTSKLEKVS KQNPTLVYPI 360
361 AIHLLGERGS LNFDRVTNTK LVEHILPLAD EQGILQLFQL LLSYVKRCPE DIASDTKAVE 420
421 WRRQWATDTM LSILRSKRSI KQEPWVRELL EIFIAYGYFE VPESEEVIPK FSEGTQNMFR 480
481 LRLMSALSYL SSSAFQQSQT DHQLGDKNWP YVALNYLLEL EKSPKNNLLI SMDESVIEIV 540
541 QKSLSVLHKV TKKIDKKAQH LQQLNAFQLL YSLVLLQVYA GDTDSIDVLE DIDNCYSKVF 600
601 NKKSKRESTS NEPTAMEILT EVMLSLLSRP SLLLRKLVDM LFTSFSEDMN RESIHLICDV 660
661 LKAKESVKDS EGMFAGEEVE EDAFGETEMD EDDFEEIDTD EIEEQSDWEM ISNQDASDNE 720
721 ELERKLDKVL EDADAKVKDE ESSEEELMND EQMLALDEKL AEVFRERKKA SNKEKKKNAQ 780
781 ETKQQIVQFK VKVIDLIDNY YKTQPNNGLG FEFLIPLLEM ILKTKHKVLE EKGQAVFRNR 840
841 LSKLKWTEEK PSSKNVLEAL KKVHVLCGKK ASLGSTGSSI SQLLLKLLAD TPYLKEGVEV 900
901 YLKSFLLWIQ EPSKSHYNAN IFHDFINWGA QQRLKHQQTS TAASSPQKTG HHENEKTNH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
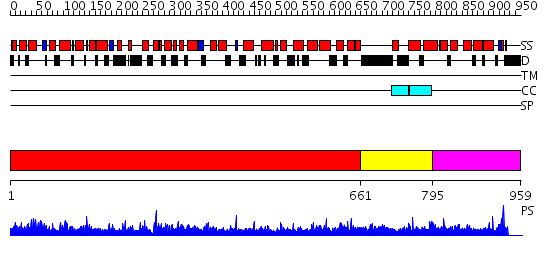
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..660] | 1.52 | Adaptin beta subunit N-terminal fragment | |
| 2 | View Details | [661..794] | 23.060997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [795..959] | 1.03599 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)