













| General Information: |
|
| Name(s) found: |
XRN2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 991 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVPALFRLL SRKFAKVITP VIEAPTEKLP DGTEIEPDLS LPNPNGVECD NLYLDMNGIV 60
61 HPCSHPEDRP APETEDEMMV AVFEYTDRIL AMVRPRQLLF IAIDGVAPRA KMNQQRSRRF 120
121 RSSREAALKE EELQAFIEEA KQQGIPIDEN ATKKKSWDSN CITPGTPFMD TLAKSLRYYI 180
181 INKLNSDPCW RNVRFILSDA SVPGEGEHKI MEFIRSQRVK PEYDPNTHHV VYGLDADLIM 240
241 LGLATHEPHF RVLREDVFFQ QGSTKKTKEE RLGIKRLDDV SETNKVPVKK PFIWLNVSIL 300
301 REYLEVELYV PNLPFPFDLE RAIDDWVFFI FFVGNDFLPH LPSLDIRDGA VERLTEIWRA 360
361 SLPHMGGYLT LDGSVNLARA EVILSAVGNQ EDDIFKRLKQ QEDRRNENYR RRQQRESNQE 420
421 SESYVDNVVI QRSVETQSTE VVTSSKSTSV DTKPPKKTQK IDAPAPVDLV NLSEKTSNRS 480
481 LGATNRELIN NRAANRLGLS REAAAVSSVN KLAASALKAQ LVSNETLQNV PLEDSIASSS 540
541 AYEDTDSIES STPVVHPIDT KVSNVGQKRK APDSTEENEN TDTVRLYEPG YRERYYEQKF 600
601 HISPDEPEKI REAVKHYVHG LCWVLLYYYQ GCPSWTWYYP YHYAPFAADF KDLASIDVKF 660
661 ELNQPFKPYE QLLGVLPAAS KNNLPEKLQT LMTDENSEII DFYPENFTID LNGKKFEWQG 720
721 VALLPFIDEN RLLNAVSKIY PQLTEEESKR NEDGSTLLFI SEHHPMFSEL VKQLYSKKRQ 780
781 GKPLKLSGKM AHGLFGKVNT NDSVIPNVSV QCPIDVTSAD ALQKYGSIDD NQSISLVFEV 840
841 PKSHFVHKSM LLRGVKMPNR VLTPEDINQV RAERSFSSRR NNGNSYRGGH QSYGVRRSYQ 900
901 SQSYSSRQSY TGVTNGFANG GVQPPWSGNG NFPRSNASYN SRGGHEGYGG RSRGGGYSNG 960
961 PPAGNHYSSN RGKGYGYQRE SYNNNNRNGY Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
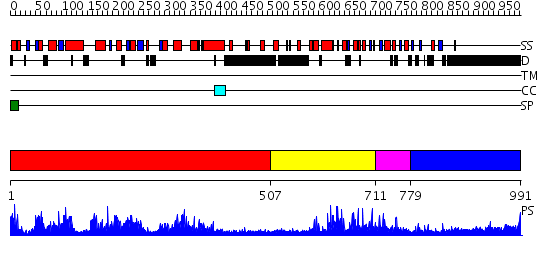
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..506] | 1.58 | Crystal structure of the human FEN1-PCNA complex | |
| 2 | View Details | [507..710] | 2.067953 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [711..778] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [779..991] | 15.30103 | No description for 1y0fB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)