













| General Information: |
|
| Name(s) found: |
gi|91212292
[NCBI NR]
gi|91073866 [NCBI NR] |
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli UTI89 |
| Length: | 410 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKVSLEKDK IKFLLVEGVH QKALESLRAA GYTNIEFHKG ALDDEQLKES IRDAHFIGLR 60
61 SRTHLTEDVI NAAEKLVAIG CFCIGTNQVD LDAAAKRGIP VFNAPFSNTR SVAELVIGEL 120
121 LLLLRGVPEA NAKAHRGVWN KLAAGSFEAR GKKLGIIGYG HIGTQLGILA ESLGMYVYFY 180
181 DIENKLPLGN ATQVQHLSDL LNMSDVVSLH VPENPSTKNM MGAKEISLMK PGSLLINASR 240
241 GTVVDIPALC DALASKHLAG AAIDVFPTEP ATNSDPFTSP LCEFDNVLLT PHIGGSTQEA 300
301 QENIGLEVAG KLIKYSDNGS TLSAVNFPEV SLPLHGGRRL MHIHENRPGV LTALNKIFAE 360
361 QGVNIAAQYL QTSAQMGYVV IDIEADEDVA EKALQAMKAI PGTIRARLLY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
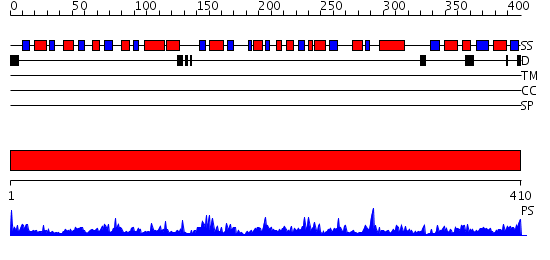
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..410] | 115.0 | The active form of phosphoglycerate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)