













| General Information: |
|
| Name(s) found: |
FBpp0292486
[FlyBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 526 amino acids |
Gene Ontology: |
|
| Cellular Component: |
subapical complex
[TAS]
cytoplasm [NAS] cell cortex [TAS] apical cortex [IDA][NAS] apical plasma membrane [NAS][TAS] |
| Biological Process: |
cell-cell junction assembly
[NAS]
germarium-derived oocyte fate determination [IMP] establishment or maintenance of cell polarity [NAS] positive regulation of neuroblast proliferation [IMP] protein amino acid phosphorylation [IEA][NAS] memory [IMP] sensory organ development [IMP] oocyte anterior/posterior axis specification [IGI] establishment or maintenance of neuroblast polarity [NAS] oocyte axis specification [TAS] synapse assembly [IMP] intracellular signaling pathway [IEA] asymmetric protein localization involved in cell fate determination [IPI] zonula adherens assembly [NAS][TAS] establishment or maintenance of epithelial cell apical/basal polarity [IMP][NAS][TAS] apical protein localization [NAS] maintenance of cell polarity [IMP] morphogenesis of a polarized epithelium [TAS] asymmetric neuroblast division [IGI] establishment or maintenance of polarity of embryonic epithelium [NAS] |
| Molecular Function: |
ATP binding
[IEA]
zinc ion binding [IEA] protein binding [IPI][ISS] protein serine/threonine kinase activity [IEA][NAS] protein kinase C activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIIWSGFCEA AQIYSTQTAT FMSGGASLFP NVPQAPGLSC DGEDRSIYRR GARRWRKLYR 60
61 VNGHIFQAKR FNRRAFCAYC QDRIWGLGRQ GFKCIQCKLL VHKKCHKLVQ KHCTDQPEPL 120
121 VKERAEESSD PIPVPLPPLP YEAMSGGAEA CETHDHAHIV APPPPEDPLE PGTQRQYSLN 180
181 DFELIRVIGR GSYAKVLMVE LRRTRRIYAM KVIKKALVTD DEDIDWVQTE KHVFETASNH 240
241 PFLVGLHSCF QTPSRLFFVI EFVRGGDLMY HMQRQRRLPE EHARFYAAEI SLALNFLHEK 300
301 GIIYRDLKLD NVLLDHEGHI KLTDYGMCKE GIRPGDTTST FCGTPNYIAP EILRGEDYGF 360
361 SVDWWALGVL LYEMLAGRSP FDLAGASENP DQNTEDYLFQ VILEKTIRIP RSLSVRAASV 420
421 LKGFLNKNPA DRLGCHRESA FMDIVSHPFF KNMDWELIAQ KEVQPPYIPN IDTGDPYVTS 480
481 NFDVQFTQEP AVLTPDDPHV IDNIDQSEFE GFEYVNPLLM SLEDCV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
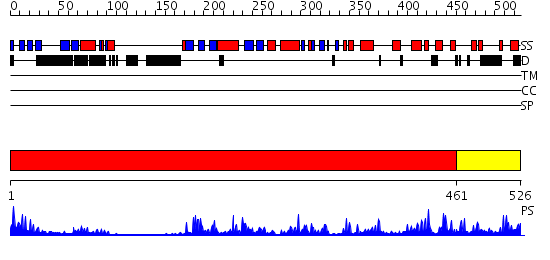
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..460] | 43.69897 | Carboxyl-terminal src kinase (csk) | |
| 2 | View Details | [461..526] | 95.154902 | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)