













| General Information: |
|
| Name(s) found: |
sdt-PH /
FBpp0110333
[FlyBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 934 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[IDA]
adherens junction [IDA] subapical complex [TAS] cell-cell adherens junction [NAS] cytoplasm [NAS] membrane [ISS][NAS] septate junction [TAS] apical cortex [IDA] plasma membrane [ISS] apical plasma membrane [IDA][NAS][TAS] |
| Biological Process: |
[NOT]establishment or maintenance of neuroblast polarity
[IEP][IMP]
cell-cell junction assembly [NAS] morphogenesis of an epithelium [NAS][TAS] zonula adherens assembly [IMP][TAS] establishment or maintenance of epithelial cell apical/basal polarity [IEP][IMP][IPI][TAS] establishment or maintenance of cell polarity [NAS] morphogenesis of a polarized epithelium [TAS] establishment or maintenance of polarity of embryonic epithelium [IMP] |
| Molecular Function: |
protein binding
[IPI][ISS][TAS]
guanylate kinase activity [ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRILKQWNRR RSGSSIVVLD GDDLKPCLPD DYISGQHHLN HQQQLQLQQQ LQQQHPLQQQ 60
61 HYRTHSGDIR EIDQEMLTML SVNQDNGPHR EMAVDCPDTF IARNKTPPRY PPPRPPQLNG 120
121 NAKPVPPPRD HLRVEKDGRL VNCSPAPQLP DRRAPGNASS GSSGATTHPL QHQQIAQIVE 180
181 PTLEQLDSIK KYQEQLRRRR EEEERIAQQN EFLRNSLRGS RKLKALQDTA TPGKAVAQQQ 240
241 QQATLATQVV GVENEAYLPD EDQPQAEQID GYGELIAALT RLQNQLSKSG LSTLAGRVSA 300
301 AHSVLASASV AHVLAARTAV LQRRRSRVSG PLHHSSLGLQ KDIVELLTQS NTAAAIELGN 360
361 LLTSHEMEGL LLAHDRIANH TDGTPSPTPT PTPAIGAATG STLSSPVAGP KRNLGMVVPP 420
421 PVVPPPLAQR GAMPLPRGES PPPVPMPPLA TMPMSMPVNL PMSACFGTLN DQNDNIRIIQ 480
481 IEKSTEPLGA TVRNEGEAVV IGRIVRGGAA EKSGLLHEGD EILEVNGQEL RGKTVNEVCA 540
541 LLGAMQGTLT FLIVPAGSPP SVGVMGGTTG SQLAGLGGAH RDTAVLHVRA HFDYDPEDDL 600
601 YIPCRELGIS FQKGDVLHVI SREDPNWWQA YREGEEDQTL AGLIPSQSFQ HQRETMKLAI 660
661 AEEAGLARSR GKDGSGSKGA TLLCARKGRK KKKKASSEAG YPLYATTAPD ETDPEEILTY 720
721 EEVALYYPRA THKRPIVLIG PPNIGRHELR QRLMADSERF SAAVPHTSRA RREGEVPGVD 780
781 YHFITRQAFE ADILARRFVE HGEYEKAYYG TSLEAIRTVV ASGKICVLNL HPQSLKLLRA 840
841 SDLKPYVVLV APPSLDKLRQ KKLRNGEPFK EEELKDIIAT ARDMEARWGH LFDMIIINND 900
901 TERAYHQLLA EINSLEREPQ WVPAQWVHNN RDES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
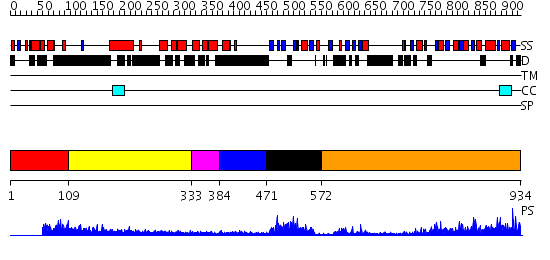
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 1.06797 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [109..332] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [333..383] | 1.52 | NMR structure of L27 heterodimer from C. elegans Lin-7 and H. sapiens Lin-2 scaffold proteins | |
| 4 | View Details | [384..470] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [471..571] | 18.154902 | Solution structure of the PDZ domain of Pals1 protein | |
| 6 | View Details | [572..934] | 56.0 | Psd-95; Guanylate kinase-like domain of Psd-95 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)