













| General Information: |
|
| Name(s) found: |
gi|109128439
[NCBI NR]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Macaca mulatta |
| Length: | 313 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGKATPPSL YSWRGVLFTC LPAARTRKRK EMSRQTATAL PTGTSKCPPS QRVPALTGTT 60
61 ASNNDLASLF ECPVCFDYVL PPILQCQSGH LVCSNCRPKL TCCPTCRGPL GSIRNLAMEK 120
121 VANSVLFPCK YASSGCEITL PHTEKADHEE LCEFRPYSCP CPGASCKWQG SLDAVMPHLM 180
181 HQHKSITTLQ GEDIVFLATD INLPGAVDWV MMQSCFGFHF MLVLEKQEKY DGHQQFFAIV 240
241 QLIGTRKQAE NFAYRLELNG HRRRLTWEAT PRSIHEGIAT AIMNSDCLVF DTSIAQLFAE 300
301 NGNLGINVTI SMC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
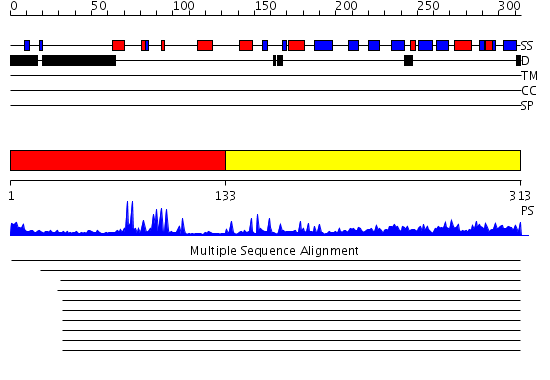
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 13.522879 | RAG1 DIMERIZATION DOMAIN | |
| 2 | View Details | [133..313] | 55.154902 | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|