













| General Information: |
|
| Name(s) found: |
RFX3
[HGNC (HUGO)]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 749 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQTSETGSDT GSTVTLQTSV ASQAAVPTQV VQQVPVQQQV QQVQTVQQVQ HVYPAQVQYV 60
61 EGSDTVYTNG AIRTTTYPYT ETQMYSQNTG GNYFDTQGSS AQVTTVVSSH SMVGTGGIQM 120
121 GVTGGQLISS SGGTYLIGNS MENSGHSVTH TTRASPATIE MAIETLQKSD GLSTHRSSLL 180
181 NSHLQWLLDN YETAEGVSLP RSTLYNHYLR HCQEHKLDPV NAASFGKLIR SIFMGLRTRR 240
241 LGTRGNSKYH YYGIRVKPDS PLNRLQEDMQ YMAMRQQPMQ QKQRYKPMQK VDGVADGFTG 300
301 SGQQTGTSVE QTVIAQSQHH QQFLDASRAL PEFGEVEISS LPDGTTFEDI KSLQSLYREH 360
361 CEAILDVVVN LQFSLIEKLW QTFWRYSPST PTDGTTITES SNLSEIESRL PKAKLITLCK 420
421 HESILKWMCN CDHGMYQALV EILIPDVLRP IPSALTQAIR NFAKSLEGWL SNAMNNIPQR 480
481 MIQTKVAAVS AFAQTLRRYT SLNHLAQAAR AVLQNTSQIN QMLSDLNRVD FANVQEQASW 540
541 VCQCDDNMVQ RLETDFKMTL QQQSTLEQWA AWLDNVMMQA LKPYEGRPSF PKAARQFLLK 600
601 WSFYSSMVIR DLTLRSAASF GSFHLIRLLY DEYMFYLVEH RVAQATGETP IAVMGEFGDL 660
661 NAVSPGNLDK DEGSEVESEM DEELDDSSEP QAKREKTELS QAFPVGCMQP VLETGVQPSL 720
721 LNPIHSEHIV TSTQTIRQCS ATGNTYTAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
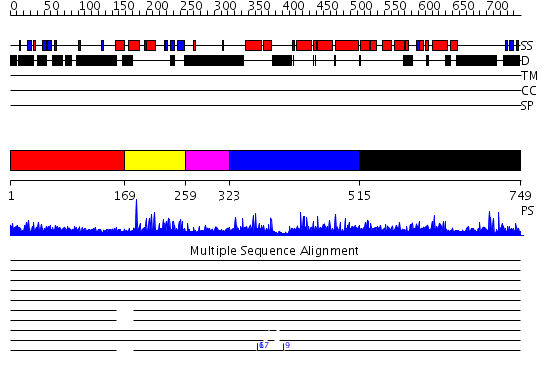
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 3.522879 | Sec24 | |
| 2 | View Details | [169..258] | 33.221849 | Class II MHC transcription factor RFX1 | |
| 3 | View Details | [259..322] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [323..514] | 2.076977 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [515..749] | 5.077965 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)