













| General Information: |
|
| Name(s) found: |
HSF2
[HGNC (HUGO)]
|
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 536 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKQSSNVPAF LSKLWTLVEE THTNEFITWS QNGQSFLVLD EQRFAKEILP KYFKHNNMAS 60
61 FVRQLNMYGF RKVVHIDSGI VKQERDGPVE FQHPYFKQGQ DDLLENIKRK VSSSKPEENK 120
121 IRQEDLTKII SSAQKVQIKQ ETIESRLSEL KSENESLWKE VSELRAKHAQ QQQVIRKIVQ 180
181 FIVTLVQNNQ LVSLKRKRPL LLNTNGAQKK NLFQHIVKEP TDNHHHKVPH SRTEGLKPRE 240
241 RISDDIIIYD VTDDNADEEN IPVIPETNED VISDPSNCSQ YPDIVIVEDD NEDEYAPVIQ 300
301 SGEQNEPARE SLSSGSDGSS PLMSSAVQLN GSSSLTSEDP VTMMDSILND NINLLGKVEL 360
361 LDYLDSIDCS LEDFQAMLSG RQFSIDPDLL VDLFTSSVQM NPTDYINNTK SENKGLETTK 420
421 NNVVQPVSEE GRKSKSKPDK QLIQYTAFPL LAFLDGNPAS SVEQASTTAS SEVLSSVDKP 480
481 IEVDELLDSS LDPEPTQSKL VRLEPLTEAE ASEATLFYLC ELAPAPLDSD MPLLDS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
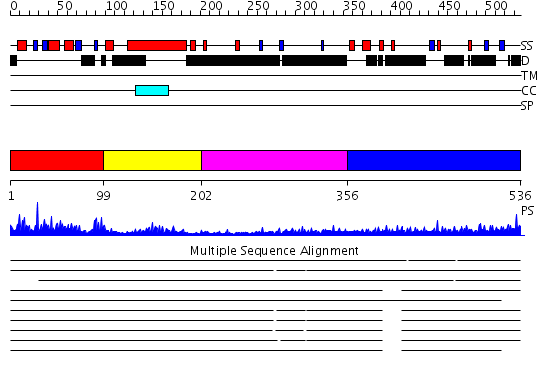
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 36.69897 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 2 | View Details | [99..201] | 8.60206 | No description for PF00447.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [202..355] | 3.198993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [356..536] | 8.156995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)