













| General Information: |
|
| Name(s) found: |
gi|108862276
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVYLHKSPFH HRGNTNISPS TTLSRSWVRP PQIRGLPCII LPPPRQSPRS AADAHMAAPS 60
61 GSGEAAPASS TYYDVYGPDA KPDVVFKEAT SNSTLNLQDV QGLVTWVIGD GMLPSWVFVK 120
121 NKPLIPKVVL LYVPGLDAAL YMSQSRHLSS LKEFCGNPKP VLASSCIPDE RHTIDALLTC 180
181 RVKRKRALKT TDQSHESDGQ EKLSSLDDLK DIPFPIKYYT LSEKDLEDNG YNFSLEGFVP 240
241 TVSAPPGSSP YAILALDCEM CVTAAGFELT RVTLVDIKGE VVLDKLVKPA NPITDYNTRF 300
301 SGITAEMLAD VTTTLQEIQE EFVGLVHKET VLVGHSLEND LMALRISHDL IIDTAVLYKH 360
361 NRGHRFKIAL RVLAKKFLNR EIQNTGSGHD SVEDAKAALE LALLKIKYGP DFGSPPSTSR 420
421 RKLASILHES GKKCSLIDDA SVLERYSDAS CNSIAVFSDD DALSRSMKEV KNDKVSFVWT 480
481 QFSKLISYLR TRAQDPDKVK SCVAEAIALQ TCDRKTAQKR KKHQTCPELK EILIGLDKKI 540
541 RKLYSILPDN AMLIICSGHG DTPLVQRLRK MLKQEEETVE SRESIVKALE DIQAQAEVAL 600
601 CFCCVKH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
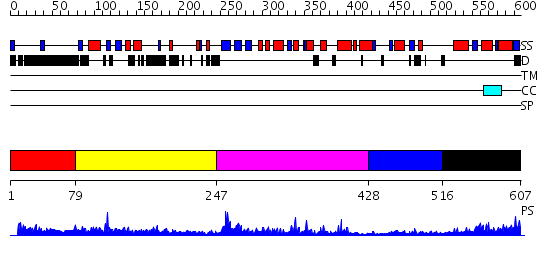
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [79..246] | 1.451989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [247..427] | 26.69897 | human ISG20 | |
| 4 | View Details | [428..515] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [516..607] | 27.375995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)