













| General Information: |
|
| Name(s) found: |
XRCC5
[HGNC (HUGO)]
|
| Description(s) found:
Found 41 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 732 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRSGNKAAV VLCMDVGFTM SNSIPGIESP FEQAKKVITM FVQRQVFAEN KDEIALVLFG 60
61 TDGTDNPLSG GDQYQNITVH RHLMLPDFDL LEDIESKIQP GSQQADFLDA LIVSMDVIQH 120
121 ETIGKKFEKR HIEIFTDLSS RFSKSQLDII IHSLKKCDIS LQFFLPFSLG KEDGSGDRGD 180
181 GPFRLGGHGP SFPLKGITEQ QKEGLEIVKM VMISLEGEDG LDEIYSFSES LRKLCVFKKI 240
241 ERHSIHWPCR LTIGSNLSIR IAAYKSILQE RVKKTWTVVD AKTLKKEDIQ KETVYCLNDD 300
301 DETEVLKEDI IQGFRYGSDI VPFSKVDEEQ MKYKSEGKCF SVLGFCKSSQ VQRRFFMGNQ 360
361 VLKVFAARDD EAAAVALSSL IHALDDLDMV AIVRYAYDKR ANPQVGVAFP HIKHNYECLV 420
421 YVQLPFMEDL RQYMFSSLKN SKKYAPTEAQ LNAVDALIDS MSLAKKDEKT DTLEDLFPTT 480
481 KIPNPRFQRL FQCLLHRALH PREPLPPIQQ HIWNMLNPPA EVTTKSQIPL SKIKTLFPLI 540
541 EAKKKDQVTA QEIFQDNHED GPTAKKLKTE QGGAHFSVSS LAEGSVTSVG SVNPAENFRV 600
601 LVKQKKASFE EASNQLINHI EQFLDTNETP YFMKSIDCIR AFREEAIKFS EEQRFNNFLK 660
661 ALQEKVEIKQ LNHFWEIVVQ DGITLITKEE ASGSSVTAEE AKKFLAPKDK PSGDTAAVFE 720
721 EGGDVDDLLD MI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC145 from april 2005 | Okada, M., et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
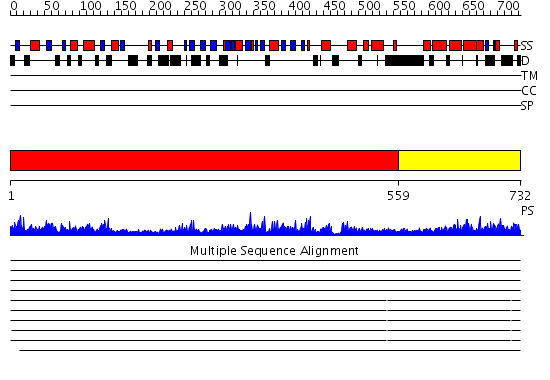
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..558] | 171.0 | Ku80 subunit | |
| 2 | View Details | [559..732] | 51.30103 | Three-dimensional structure of Ku80 CTD |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)