













| General Information: |
|
| Name(s) found: |
NUDT12
[HGNC (HUGO)]
|
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 462 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVKRSLKQ EIVTQFHCSA AEGDIAKLTG ILSHSPSLLN ETSENGWTAL MYAARNGHPE 60
61 IVQFLLEKGC DRSIVNKSRQ TALDIAVFWG YKHIANLLAT AKGGKKPWFL TNEVEECENY 120
121 FSKTLLDRKS EKRNNSDWLL AKESHPATVF ILFSDLNPLV TLGGNKESFQ QPEVRLCQLN 180
181 YTDIKDYLAQ PEKITLIFLG VELEIKDKLL NYAGEVPREE EDGLVAWFAL GIDPIAAEEF 240
241 KQRHENCYFL HPPMPALLQL KEKEAGVVAQ ARSVLAWHSR YKFCPTCGNA TKIEEGGYKR 300
301 LCLKEDCPSL NGVHNTSYPR VDPVVIMQVI HPDGTKCLLG RQKRFPPGMF TCLAGFIEPG 360
361 ETIEDAVRRE VEEESGVKVG HVQYVACQPW PMPSSLMIGC LALAVSTEIK VDKNEIEDAR 420
421 WFTREQVLDV LTKGKQQAFF VPPSRAIAHQ LIKHWIRINP NL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
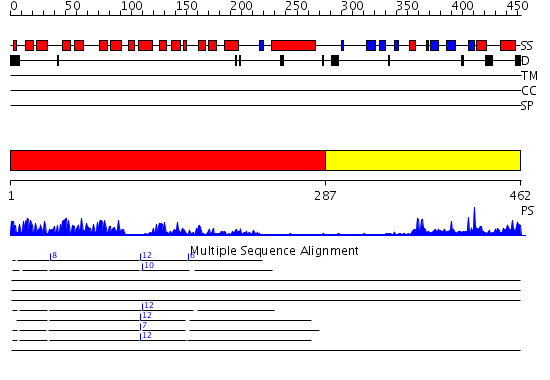
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..286] | 46.0 | Ankyrin-R | |
| 2 | View Details | [287..462] | 16.0 | Crystal structure of NADH pyrophosphatase (1790429) from Escherichia coli k12 at 2.20 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)