













| General Information: |
|
| Name(s) found: |
CHR25_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 910 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
DNA repair
[ISS]
double-strand break repair via homologous recombination [IMP] response to gamma radiation [IEP] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA][ISS] DNA binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEEDEEILS SSDCDDSSDS YKDDSQDSEG ENDNPECEDL AVVSLSSDAD RKSKNVKDLL 60
61 RGNLVVQRQP LLPRVLSVSD GAAVCRKPFK PPCSHGYDST GQLSRRLSAR KRFVPWGSST 120
121 PVVVALPTKL EASTNIERDE EEEVVCLPPD IEPLVLWQSE EDGMSNVTTI MVHSVLVKFL 180
181 RPHQREGVQF MFDCVSGLHG SANINGCILA DDMGLGKTLQ SITLLYTLLC QGFDGTPMVK 240
241 KAIIVTPTSL VSNWEAEIKK WVGDRIQLIA LCESTRDDVL SGIDSFTRPR SALQVLIISY 300
301 ETFRMHSSKF CQSESCDLLI CDEAHRLKND QTLTNRALAS LTCKRRVLLS GTPMQNDLEE 360
361 FFAMVNFTNP GSLGDAAHFR HYYEAPIICG REPTATEEEK NLAADRSAEL SSKVNQFILR 420
421 RTNALLSNHL PPKIIEVVCC KMTTLQSTLY NHFISSKNLK RALADNAKQT KVLAYITALK 480
481 KLCNHPKLIY DTIKSGNPGT VGFENCLEFF PAEMFSGRSG AWTGGDGAWV ELSGKMHVLS 540
541 RLLANLRRKT DDRIVLVSNY TQTLDLFAQL CRERRYPFLR LDGSTTISKR QKLVNRLNDP 600
601 TKDEFAFLLS SKAGGCGLNL IGANRLVLFD PDWNPANDKQ AAARVWRDGQ KKRVYVYRFL 660
661 STGTIEEKVY QRQMSKEGLQ KVIQHEQTDN STRQGNLLST EDLRDLFSFH GDVRSEIHEK 720
721 MSCSRCQNDA SGTENIEEGN ENNVDDNACQ IDQEDIGGFA KDAGCFNLLK NSERQVGTPL 780
781 EEDLGSWGHH FTSKSVPDAI LQASAGDEVT FVFTNQVDGK LVPIESNVSP KTVESEEHNR 840
841 NQPVNKRAFN KPQQRPREPL QPLSLNETTK RVKLSTYKRL HGNSNIDDAQ IKMSLQRPNL 900
901 VSVNHDDDFV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
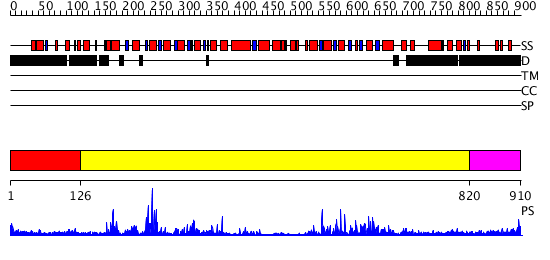
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..125] | 9.0 | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | |
| 2 | View Details | [126..819] | 104.0 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 3 | View Details | [820..910] | 4.0 | Crystal structure of SecA:ADP in an open conformation from Bacillus Subtilis |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)