













| General Information: |
|
| Name(s) found: |
SMN2
[HGNC (HUGO)]
SMN1 [HGNC (HUGO)] |
| Description(s) found:
Found 54 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 294 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMSSGGSGG GVPEQEDSVL FRRGTGQSDD SDIWDDTALI KAYDKAVASF KHALKNGDIC 60
61 ETSGKPKTTP KRKPAKKNKS QKKNTAASLQ QWKVGDKCSA IWSEDGCIYP ATIASIDFKR 120
121 ETCVVVYTGY GNREEQNLSD LLSPICEVAN NIEQNAQENE NESQVSTDES ENSRSPGNKS 180
181 DNIKPKSAPW NSFLPPPPPM PGPRLGPGKP GLKFNGPPPP PPPPPPHLLS CWLPPFPSGP 240
241 PIIPPPPPIC PDSLDDADAL GSMLISWYMS GYHTGYYMGF RQNQKEGRCS HSLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
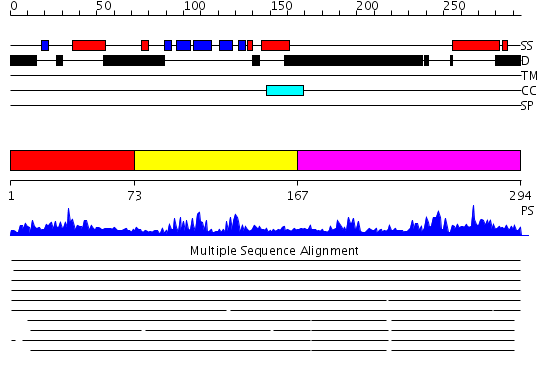
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] | 1.135974 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [73..166] | 28.30103 | Survival motor neuron protein 1, smn | |
| 3 | View Details | [167..294] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)