













| General Information: |
|
| Name(s) found: |
PRKCE
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 737 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVFNGLLKI KICEAVSLKP TAWSLRHAVG PRPQTFLLDP YIALNVDDSR IGQTATKQKT 60
61 NSPAWHDEFV TDVCNGRKIE LAVFHDAPIG YDDFVANCTI QFEELLQNGS RHFEDWIDLE 120
121 PEGRVYVIID LSGSSGEAPK DNEERVFRER MRPRKRQGAV RRRVHQVNGH KFMATYLRQP 180
181 TYCSHCRDFI WGVIGKQGYQ CQVCTCVVHK RCHELIITKC AGLKKQETPD QVGSQRFSVN 240
241 MPHKFGIHNY KVPTFCDHCG SLLWGLLRQG LQCKVCKMNV HRRCETNVAP NCGVDARGIA 300
301 KVLADLGVTP DKITNSGQRR KKLIAGAESP QPASGSSPSE EDRSKSAPTS PCDQEIKELE 360
361 NNIRKALSFD NRGEEHRAAS SPDGQLMSPG ENGEVRQGQA KRLGLDEFNF IKVLGKGSFG 420
421 KVMLAELKGK DEVYAVKVLK KDVILQDDDV DCTMTEKRIL ALARKHPYLT QLYCCFQTKD 480
481 RLFFVMEYVN GGDLMFQIQR SRKFDEPRSR FYAAEVTSAL MFLHQHGVIY RDLKLDNILL 540
541 DAEGHCKLAD FGMCKEGILN GVTTTTFCGT PDYIAPEILQ ELEYGPSVDW WALGVLMYEM 600
601 MAGQPPFEAD NEDDLFESIL HDDVLYPVWL SKEAVSILKA FMTKNPHKRL GCVASQNGED 660
661 AIKQHPFFKE IDWVLLEQKK IKPPFKPRIK TKRDVNNFDQ DFTREEPVLT LVDEAIVKQI 720
721 NQEEFKGFSY FGEDLMP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
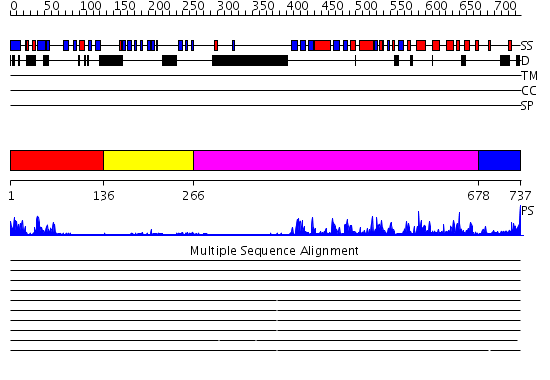
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 12.30103 | Domain from protein kinase C epsilon | |
| 2 | View Details | [136..265] | 3.22 | Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta | |
| 3 | View Details | [266..677] | 47.0 | No description for 2ozoA was found. | |
| 4 | View Details | [678..737] | 82.0 | No description for 2i0eA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)