













| General Information: |
|
| Name(s) found: |
CCT3
[HGNC (HUGO)]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 544 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGHRPVLVLS QNTKRESGRK VQSGNINAAK TIADIIRTCL GPKSMMKMLL DPMGGIVMTN 60
61 DGNAILREIQ VQHPAAKSMI EISRTQDEEV GDGTTSVIIL AGEMLSVAEH FLEQQMHPTV 120
121 VISAYRKALD DMISTLKKIS IPVDISDSDM MLNIINSSIT TKAISRWSSL ACNIALDAVK 180
181 MVQFEENGRK EIDIKKYARV EKIPGGIIED SCVLRGVMIN KDVTHPRMRR YIKNPRIVLL 240
241 DSSLEYKKGE SQTDIEITRE EDFTRILQME EEYIQQLCED IIQLKPDVVI TEKGISDLAQ 300
301 HYLMRANITA IRRVRKTDNN RIARACGARI VSRPEELRED DVGTGAGLLE IKKIGDEYFT 360
361 FITDCKDPKA CTILLRGASK EILSEVERNL QDAMQVCRNV LLDPQLVPGG GASEMAVAHA 420
421 LTEKSKAMTG VEQWPYRAVA QALEVIPRTL IQNCGASTIR LLTSLRAKHT QENCETWGVN 480
481 GETGTLVDMK ELGIWEPLAV KLQTYKTAVE TAVLLLRIDD IVSGHKKKGD DQSRQGGAPD 540
541 AGQE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
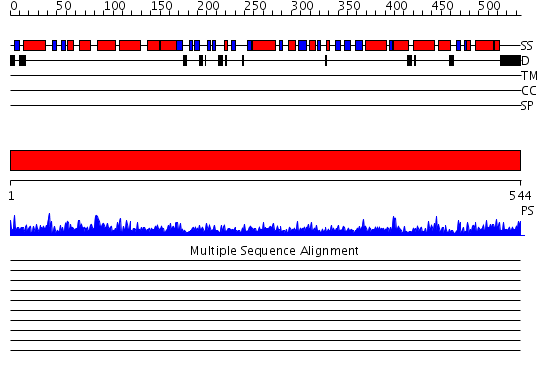
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..544] | 149.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)