













| General Information: |
|
| Name(s) found: |
DPB2 /
YPR175W
[SGD]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 692 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[TAS nucleus [IDA cytoplasm [IDA epsilon DNA polymerase complex [IDA |
| Biological Process: |
lagging strand elongation
[TAS leading strand elongation [TAS nucleotide-excision repair [TAS mismatch repair [TAS |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCEMFGSGNV LPVKIQPPLL RPLAYRVLSR KYGLSIKSDG LSALAEFVGT NIGANWRQGP 60
61 ATIKFLEQFA AVWKQQERGL FIDQSGVKEV IQEMKEREKV EWSHEHPIQH EENILGRTDD 120
121 DENNSDDEMP IAADSSLQNV SLSSPMRQPT ERDEYKQPFK PESSKALDWR DYFKVINASQ 180
181 QQRFSYNPHK MQFIFVPNKK QNGLGGIAGF LPDIEDKVQM FLTRYYLTND RVMRNENFQN 240
241 SDMFNPLSSM VSLQNELSNT NRQQQSSSMS ITPIKNLLGR DAQNFLLLGL LNKNFKGNWS 300
301 LEDPSGSVEI DISQTIPTQG HYYVPGCMVL VEGIYYSVGN KFHVTSMTLP PGERREITLE 360
361 TIGNLDLLGI HGISNNNFIA RLDKDLKIRL HLLEKELTDH KFVILGANLF LDDLKIMTAL 420
421 SKILQKLNDD PPTLLIWQGS FTSVPVFASM SSRNISSSTQ FKNNFDALAT LLSRFDNLTE 480
481 NTTMIFIPGP NDLWGSMVSL GASGTLPQDP IPSAFTKKIN KVCKNVVWSS NPTRIAYLSQ 540
541 EIVIFRDDLS GRFKRHRLEF PFNESEDVYT ENDNMMSKDT DIVPIDELVK EPDQLPQKVQ 600
601 ETRKLVKTIL DQGHLSPFLD SLRPISWDLD HTLTLCPIPS TMVLCDTTSA QFDLTYNGCK 660
661 VINPGSFIHN RRARYMEYVP SSKKTIQEEI YI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
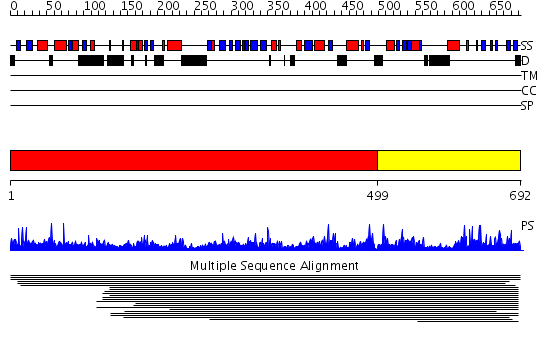
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..498] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [499..692] | 1.015968 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)